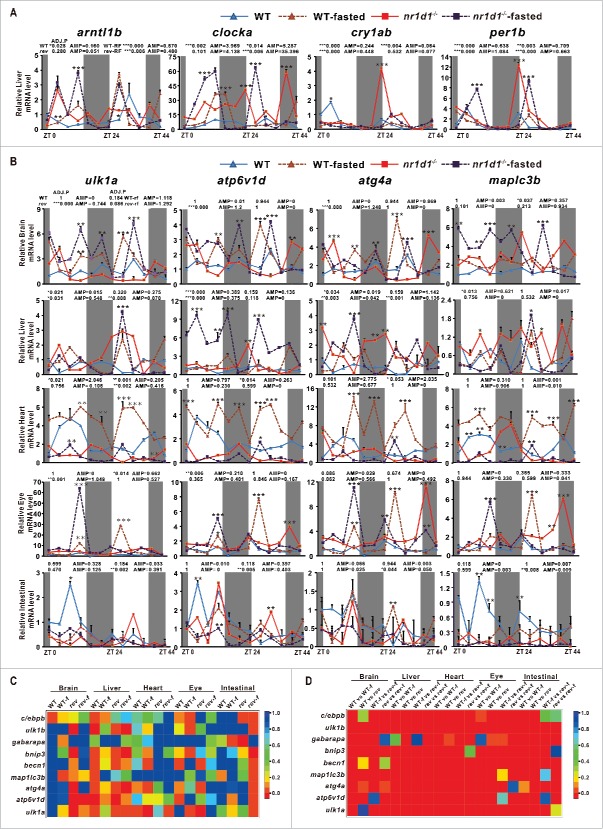
Figure 7.
Effects of fasting and loss of Nr1d1 on expression of autophagy genes and circadian clock genes. (A) qRT-PCR analysis of core circadian clock genes in the liver of nr1d1 mutant (fed), fasted nr1d1 mutant, wild-type (fed) and fasted wild-type fish. (B) qRT-PCR analysis of autophagy genes map1lc3b, atp6v1d, ulk1a and atg4a in the adult peripheral organs of nr1d1 mutant, fasted nr1d1 mutant, fasted wild-type and wild-type fish under LD. Following 2-wk fasting, 4-mo-old wild-type and nr1d1 mutant fish, and their controls were sacrificed, and the brains, the livers, the hearts, the eyes, and the intestines were dissected out and collected for RNA extraction at 12 time points with 4-h interval under LD for a total of consecutive 2 d. Each sample contains organs from at least 5 male fishes. Three independent sets of samples were used. The mRNA expression pattern was analysis by JTK-CYCLE method. ADJ.P for adjusted minimal P values (*, P ≤ 0 .05; **, P ≤ 0 .01; ***, P ≤ 0 .001), AMP for amplitude. Two-way ANOVA with the Tukey post hoc test was conducted (*, P≤0 .05; **, P ≤ 0 .01; ***, P ≤ 0 .001). (C) Heatmap comparing ADJ.P values of autophagy and autophagy-related genes in different zebrafish organs of wild-type (fed), wild-type-fasted, nr1d1 (fed) and nr1d1 -fasted groups. (D) Heatmap showing correlation coefficiencies (R) of P values from 2-way ANOVA of autophagy and autophagy-related genes in different zebrafish organs in terms of wild-type (fed) vs wild-type-fasted, wild-type (fed) vs nr1d1 (fed), wild-type-fasted vs nr1d1-fasted, and nr1d1 (fed) vs nr1d1-fasted. WT-f, WT-fasted.