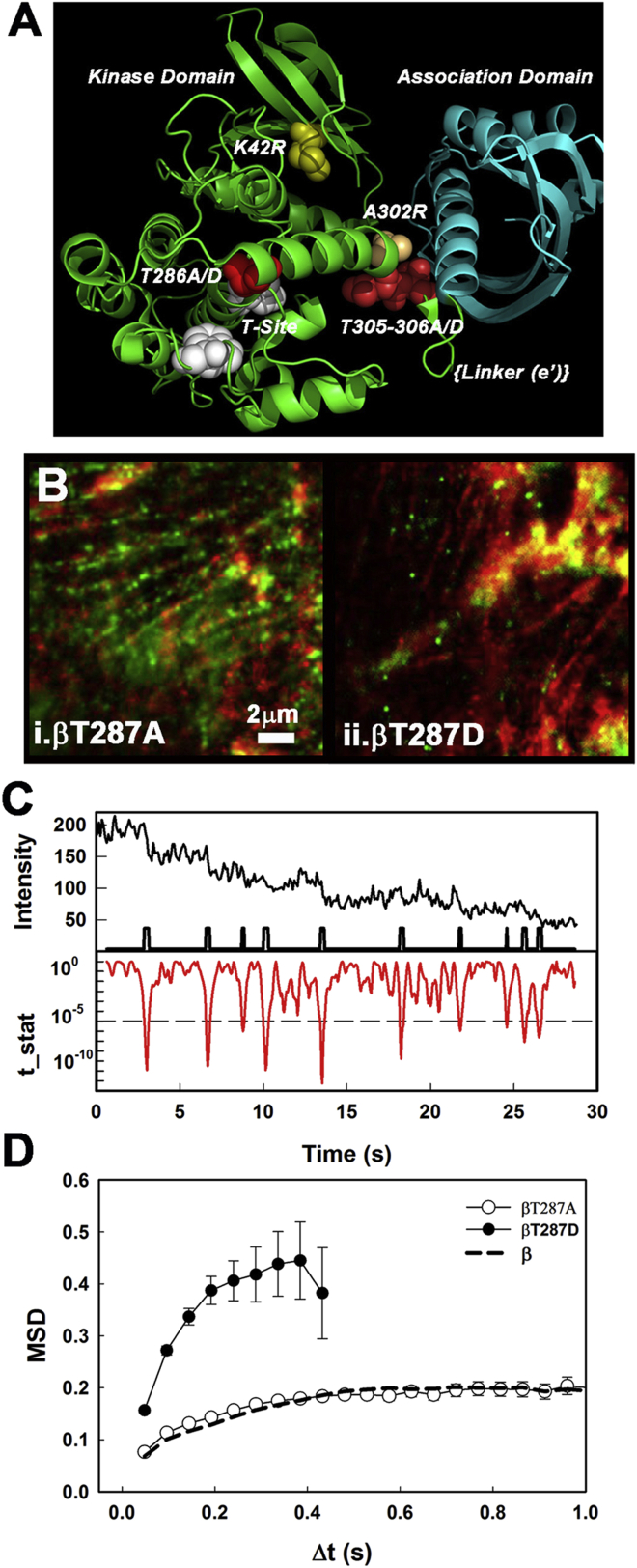
Figure 5.
The T287D point mutation downregulates association with the actin cytoskeleton. (A) Atomic structure (PDB: 3SOA) of rat CaMKII (18). The positions of the mutated sites (K42 (yellow), T286 (red), A302 (peach), and T305/T306 (magenta)) studied; the junction of kinase (green) and association (blue) domains where the splice E′ linker segment would be located; and the substrate-binding T-site (white) are shown in relation to the secondary-structure elements (cartoon representation). (B) Superimposed averaged images, processed as in Fig. 3A, show localization of single molecules (Fig. S4; Movies S2 and S3) of (i) the dephosphorylated mimic, β287A (550 frames, Ppix = 0.29, Prand = 0.16 ± 0.07, n = 6511) and (ii) the phosphomimic, β287D (475 frames (Ppix = 0.08, Prand = 0.0 ± 0.08, n = 1020) with tRFP-actin (red, 100 frames). The mean tRFP intensities were 105 ± 6 (βT287A) and 213 ± 75 (βT287D) counts/pixel. (C) Photobleaching profile of a long-lived β287A-GFP track (upper black line plot) with the corresponding t-test statistic (t-stat) based on a rolling, nonoverlapping 12-frame window (lower red line plot). The t-stat axis denotes the probability that successive 12-frame segments have the same mean. The probability threshold was set to 10−5 for detection of a step change. The bars on the time axis of the upper plot mark the 10 steps identified by the t-test. The mean lifetime and intensity decrease per step were 2.6 ± 0.4 s and 15.5 counts/pixel, respectively. (D) Average track MSD-versus-Δt plots for the β287D and β287A populations. The dashed line is the plot for the native β population redrawn from Fig. 3C.