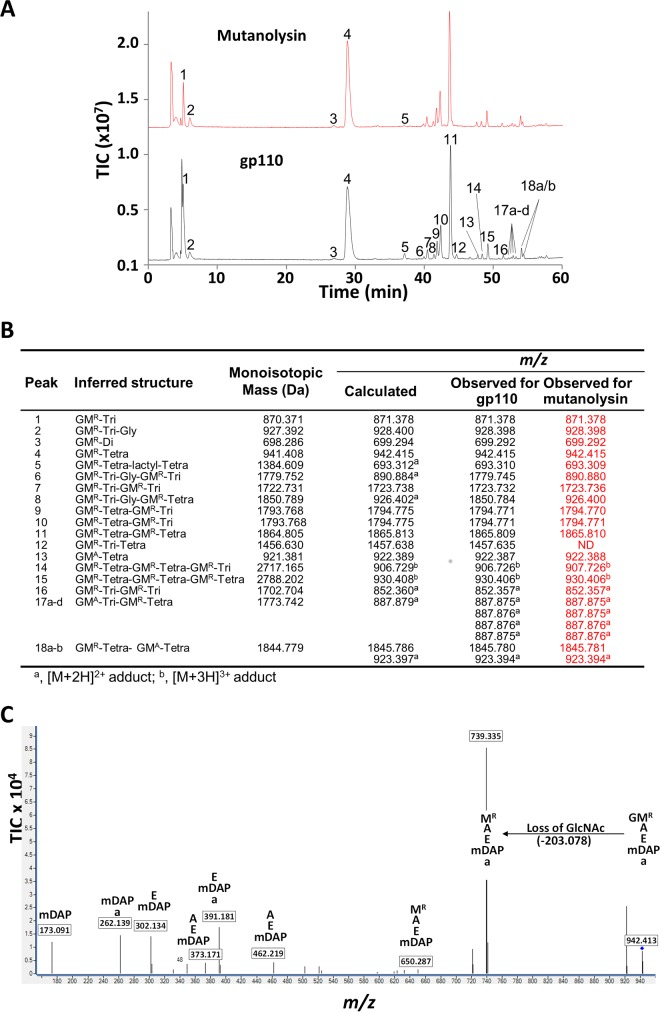
FIG 4.
Determination of Gp110 cleavage specificity. (A) LC-MS analysis of E. coli BW25113 Δlpp peptidoglycan digested by mutanolysin and recombinant Gp110. Soluble muropeptides were reduced and analyzed by RP-HPLC coupled to MS. Peaks corresponding to m/z values matching previously identified muropeptides are numbered. (B) Inferred structures, theoretical monoisotopic masses, and theoretical and observed m/z values of the peaks identified in panel A. Di, l-Ala-d-Glu; Tri, l-Ala-d-Glu-m-DAP; Tetra, l-Ala-d-Glu-m-DAP-d-Ala. (C) LC-MS/MS analysis of the major disaccharide peptide (peak 4) solubilized by Gp110. The fragmentation pattern of the [M + H]+ ion at m/z 942.414 was typical of a disaccharide tetrapeptide (DS-Tetra). The fragmentation event leading to the loss of a nonreduced GlcNAc residue (203.078) indicates that Gp110 displays N-acetylmuramidase (lysozyme) activity. The sequence of peptide fragments is indicated above their respective m/z values (boxed). A, l-Ala or d-Ala; a, C-terminal d-Ala; m-DAP, meso-diaminopimelic acid; E, γ-d-Glu; MR, reduced MurNAc; G, GlcNAc. TIC, total ion count.