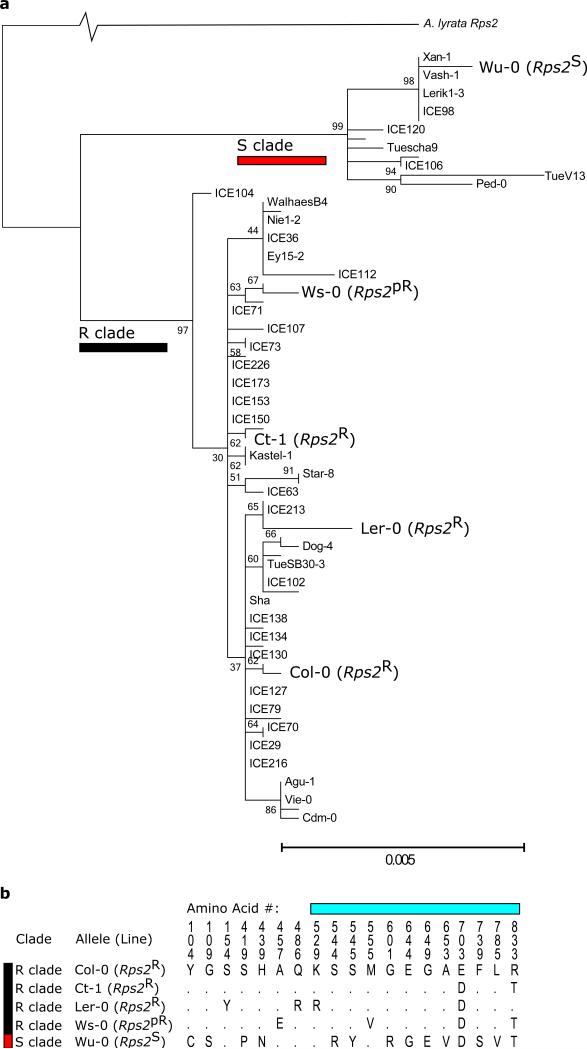
Figure 1.
Natural variation in Rps2 captured by the transgenic allelic series. Red and black bars indicate the susceptible and resistant clades of Rps2 alleles, respectively. a) The two clades of Rps2 alleles were inferred from the coding sequence of Rps2 using maximum likelihood, and included 80 genomes from Cao et al.37 and Sanger sequencing of the five alleles used in this study. The percentage of trees in which the associated taxa clustered together out of 1000 bootstrap replicates is shown next to the branches. b) Amino acid variation in the alleles used in this study. Cyan bar indicates the Leucine-rich repeat region of Rps2; Rps2R and Rps2pR lines are resistant and partially resistant to Pseudomonas syringae pv. avrRpt2.