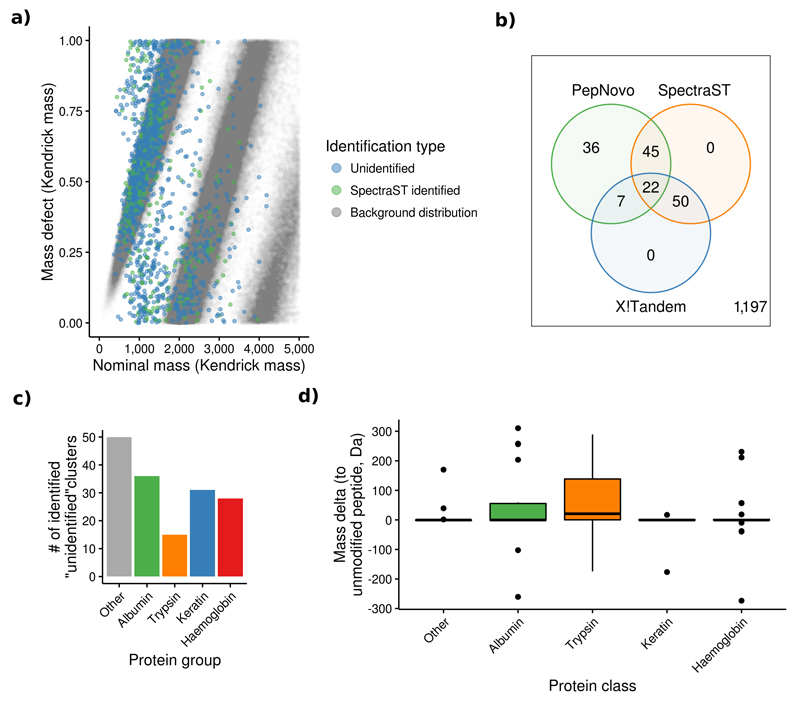
Figure 4.
Overview of the results of the analysis of clusters containing only unidentified spectra. (a) The mass defect analysis showed that ~80% of the unidentified human spectra have a similar distribution as the background one created from all in silico digested tryptic peptides in UniProtKB/SwissProt. The remaining 20% of spectra not included within this distribution may be explained by the fact that only unmodified, fully tryptic peptides were considered for this distribution. (b) 160 (12%) of the large unidentified human clusters were identified using SpectraST, X!Tandem and PepNovo. (c) More than 50% of these identifications were peptides coming from albumin, trypsin, keratin and haemoglobin. (d) Only trypsin peptides were commonly modified (e.g. dimethylated, center line marks the median, edges the first and third quartile, whiskers extend to +/-1.58 times the inter-quartile ratio divided by the square root of the number of observations, single points denote measurements outside this range).