Fig. 1.
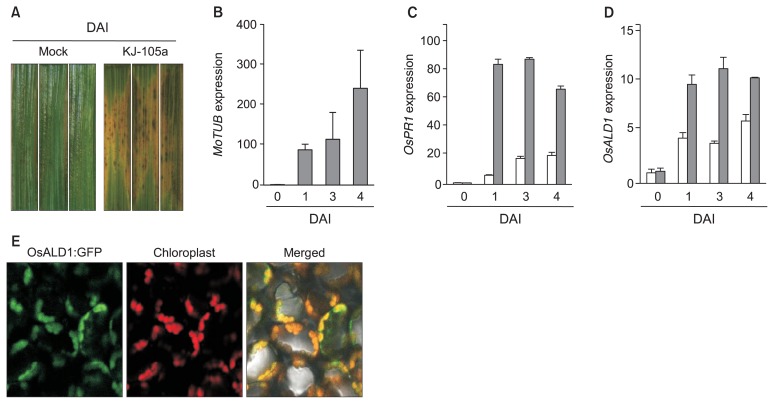
An OsALD1 gene, whose products accumulated at chloroplast, was strongly expressed in the infected leaves of rice plants with rice blast fungus. (A) Symptom development of rice blast disease in Oryza sativa cv. Dongjin infected by a Magnaporthe oryzae KJ-105a isolate. (B) Fungal growth was verified by quantifying expression of M. oryzae β-Tubulin2 (MoTUB). The relative expression level was calculated by a 2−ΔΔCT method (Livak and Schmittgen, 2001). A rice ubiquitin gene was used as an internal reference gene. (C, D) Infection with the rice blast fungus triggered expression of O. sativa PATHOGENESIS-RELATED PROTEIN1b (OsPR1b) (C) and OsALD1 (D) genes in the infected leaf tissues. Relative expression ratios were computed by a standard curve-based method (Pfaffl, 2001). mRNA levels of each sample were normalized by that of cv. Dongjin plants before mock-inoculation. Data represent the average with standard deviation (n = 3). Either fungal spore suspension (gray bars) (5 × 105 conidia/ml) or water (white bars) was inoculated with a paintbrush on the leaves (A–D). These experiments were repeated twice with the same results. (E) OsALD1 proteins localized at chloroplast in the leaves of Nicotiana benthamiana. OsALD1:GFP construct, whose expression was conditionally controlled by dexamethasone (DEX)-inducible promoter, was introduced in the leaves of N. benthamiana in accordance with an Agrobacterium-mediated transient expression protocol. Green fluorescence protein (GFP) was visualized 1 day after DEX (30 μM) treatment under a confocal microscopy (× 100). DAI, days after inoculation.