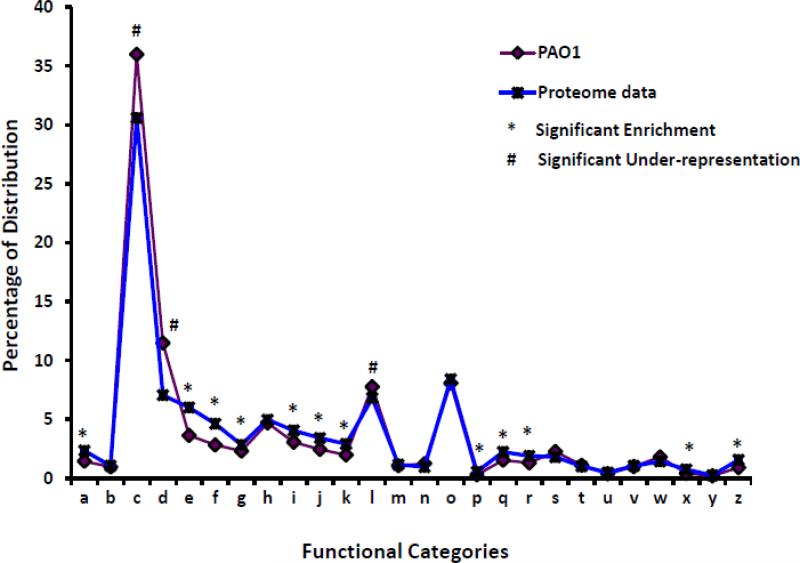
Fig. 2. Functional categorization of the total proteins identified in GeLC-MS analysis by PseudoCAP (winsor 2011).
All the non-redundant proteins (2965) identified by crux and Inspect were functionally categorized and plotted as percentage distribution of each category in PAO1 (purple) vs. proteome data (blue). Hypergeometric analysis was performed to determine significantly enriched (*) or under-represented (#) categories (p-values of ≤ 0.002). The functional categories are (a) DNA replication, recombination, modification and repair; (b) fatty acid and phospholipid metabolism; (c) hypothetical; (d) membrane proteins; (e) amino acid biosynthesis, metabolism; (f) translation, post-translational modification, degradation; (g) cell wall/LPS/capsule; (h) transport of small molecules; (i) energy metabolism; (j) biosynthesis of cofactors, prosthetic groups, carriers; (k) adaptation, protection; (l) transcriptional regulators; (m) two-component regulatory systems; (n) secreted factors- toxins, enzymes, alginate; (o) putative enzymes; (p) chaperones, heat-shock proteins; (q) central intermediary metabolism; (r) nucleotide biosynthesis and metabolism; (s) carbon compound catabolism; (t) motility and attachment; (u) chemotaxis; (v) related to phage, transposon, plasmid; (w) protein secretion, export apparatus; (x) cell division; (z) transcription, RNA processing, degradation.