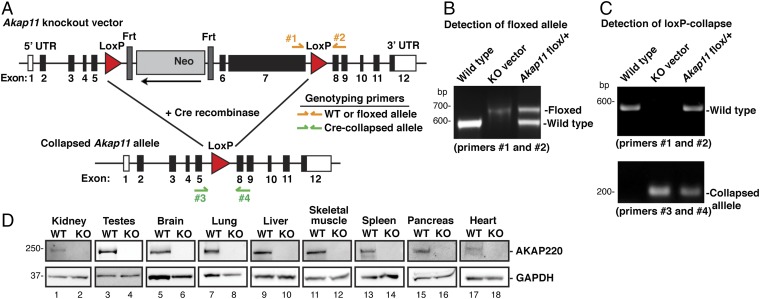
Fig. 2.
Production and verification of the AKAP220-KO mouse model. (A) Schematic of AKAP220-KO vector shows LoxP sites flanking exons 6 and 7 of Akap11, the gene encoding AKAP220 (83% of coding region). An Frt-flanked neomycin (Neo) cassette and a thymidine kinase (TK) cassette were inserted as positive and negative selection markers. Genotyping primers were designed to differentiate between WT and floxed alleles (primers 1 and 2), and a separate primer pair was designed to detect the collapsed allele (primers 3 and 4). (B) PCR analysis with genotyping primers 1 and 2 detected the WT allele and the floxed Akap11 allele that has a 78-bp insertion. WT DNA and purified Akap11-KO vector DNA were used as positive controls. (C) Floxed AKAP220 mice were crossed with mice expressing EIIa-Cre to generate global AKAP220-KO mice. PCR using genotyping primers 3 and 4 detected the collapsed Akap11 allele. Two separate PCR reactions using primers 1 and 2 and primers 3 and 4 determined genotype of each animal. (D) Western blots show AKAP220 protein expression. (Top) Tissue panel from WT and AKAP220-KO mice. (Bottom) GAPDH was used as a loading control.