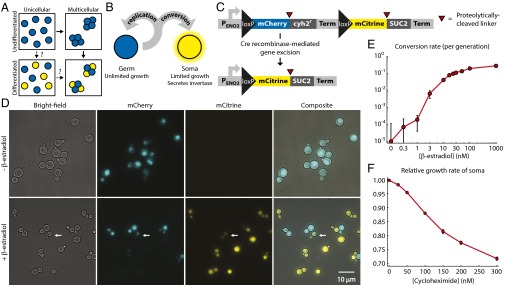
Fig. 1.
Engineering yeast as a model for somatic differentiation. (A) Alternative evolutionary trajectories for the evolution of development. Multicellularity and somatic differentiation have separate biological bases and thus probably evolved sequentially. In some clades, the persistence of likely evolutionary intermediates suggests that multicellularity arose first (solid arrows); no examples of evolution through a unicellular differentiating intermediate (dashed arrows) are known. Somatic cells are colored yellow. (B) Schematic of germ-soma division of labor model engineered in our differentiating strains. (C) Cell type-defining locus in the differentiating strains. Each pair of cell type-specific proteins (mCherry and Cyh2r for germ cells, mCitrine and Suc2 for somatic cells) is initially expressed as a single polypeptide with a ubiquitin linker (red triangles). Cellular deubiquitinating enzymes cleave this linker posttranslationally at its C terminus (34), allowing the two resulting peptides to localize and function independently: for example, Suc2 enters the secretory pathway, whereas mCitrine remains in the cytoplasm. A transcriptional terminator (Term) blocks expression of the somatic cell proteins in germ cells. Cre recombinase-mediated gene excision between loxP sites removes the terminator, as well as the germ cell-specific genes. (D) The unicellular differentiating strain (yMEW192) during growth in yeast extract-peptone-dextrose media (YPD) before (Top) or 5 h after addition of 1 μM β-estradiol (Bottom). The white arrow indicates a budded cell that has recently undergone conversion and contains both mCherry and mCitrine proteins and thus fluoresces in both channels (cf Fig. S1). (E) Conversion rates estimated by flow cytometry during growth in YPD containing β-estradiol. Error bars represent 95% confidence intervals (CIs) of the mean, determined using data obtained from three biological replicates. (F) The growth disadvantage of somatic cells relative to germ cells was measured by growing them in coculture in YPD plus cycloheximide and determining the change in the ratio between cell types over multiple rounds of cell division. Error bars represent 95% CIs of the mean, determined using data obtained from three biological replicates.