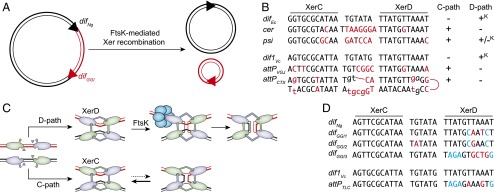
Fig. 1.
(A) Schematic of XerCD-mediated excision of the GGI. Black double lines, N. gonorrheae chromosomal DNA; red double lines, GGI DNA; black triangles, difNg and red triangle, difGGI. (B) Sequence alignment of the Xer recombination site of mobile elements and their cognate partner chromosomal dif site; attPCTX, CTXϕ attachment site; attPVGJ, VGJϕ attachment site; cer, core dimer resolution site of plasmid ColE1; difEc, E. coli dif; dif1Vc, V. cholerae chr1 dif; and psi, core dimer resolution site of plasmid pSC101. Apart from attPCTX, which is the stem of a forked hairpin from the single-stranded form of the genome of CTXϕ, a single of the two DNA strands is represented in the 5′ to 3′ orientation from left to right. Bases of cer and psi that differ from difEc and bases from attPCTX and attPVGJ that differ from dif1Vc are indicated in red. Plus (+) and minus (–) signs indicate whether the sites can engage in recombination pathways initiated by XerC or XerD strand exchanges; +K denotes FtsK-dependent recombination pathways. (C) Schematic of Xer recombination. XerD and XerC are represented in magenta and green, respectively. Following the Cre paradigm, the active pair of recombinases are drawn with their extreme C-terminal domains contacting the partner recombinases in cis. Blue circles represent the hexamer of FtsK. (D) Sequence alignment of difNg, the three different types of difGGI, dif1Vc, and attPTLC. Bases of the dif-like site of mobile elements that differ from their cognate dif partner are highlighted in color, with blue highlighting those that are identical in difGGI1 and difGGI2 and in attPGGI3 and attPTLC.