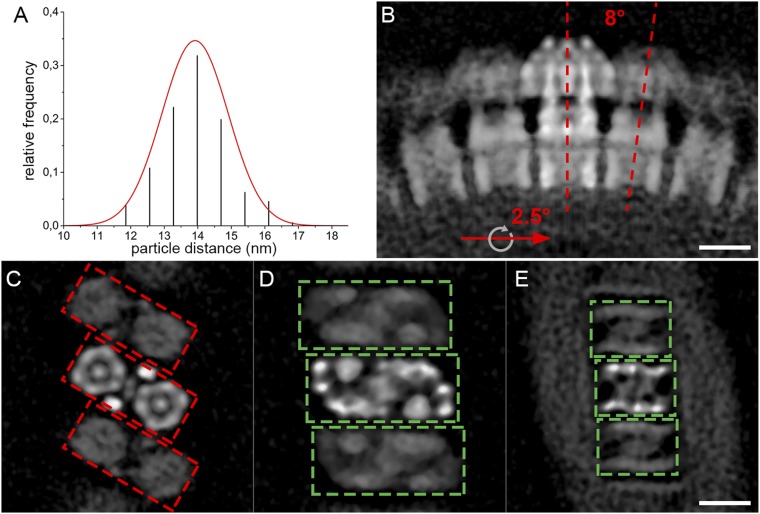
Fig. S6.
Macromolecular organization of the mitochondrial ATP synthase in P. tetraurelia. (A) Center-to-center distances between neighboring ATP synthase dimers (n = 176) in mitochondrial membranes. The mean distance, fitted with a Gaussian distribution, is 14 ± 1 nm (SD). (B) Side view and (C–E) cross-sections through the subtomogram average showing the average position of neighboring dimers. Neighboring dimers are related by a 2.5° rotation around the axis of the row and an 8° rotation around a perpendicular axis (red lines, B). (C) F1 heads, (D) central stalk and basal end of peripheral stalk, and (E) intracrista domain. The 30° offset in the positioning of the F1 subcomplexes (red boxes, C) relative to the intracrista region (green boxes, D and E) in the U-shaped dimer results in a zigzag arrangement of the catalytic domain, whereas the intracrista region forms a ribbed array. The weaker density of the peripheral dimers relative to the central dimer reflects the positional variability of ATP synthase dimers within the helical arrays. (Scale bars, 10 nm.)