Significance
Degeneration of midbrain dopamine neurons is the main pathological hallmark of Parkinson’s disease. Identifying transcriptional programs that maintain these neurons in the adult brain will help us understand their specific vulnerability. Here, we show that the survival of dopaminergic neurons requires the ongoing action of LIM-homeodomain transcription factors Lmx1a and Lmx1b. We discovered an Lmx1a/b-dependent pathway maintaining mitochondrial functions in midbrain dopaminergic neurons. Accordingly, ablation of Lmx1a/b results in impaired respiratory chain activity, increased oxidative stress, and mitochondrial DNA damage and causes Lewy neurite-like pathology. Importantly, deletion of Lmx1a/b links metabolic impairment, α-synuclein inclusions, and progressive neuronal loss. Modulation of this pathway opens new strategies to slow down or prevent the death of vulnerable neurons in Parkinson’s disease.
Keywords: transcription factors, Parkinson's disease, mitochondrial dysfunctions, protein aggregates, dopamine neurons
Abstract
The LIM-homeodomain transcription factors Lmx1a and Lmx1b play critical roles during the development of midbrain dopaminergic progenitors, but their functions in the adult brain remain poorly understood. We show here that sustained expression of Lmx1a and Lmx1b is required for the survival of adult midbrain dopaminergic neurons. Strikingly, inactivation of Lmx1a and Lmx1b recreates cellular features observed in Parkinson’s disease. We found that Lmx1a/b control the expression of key genes involved in mitochondrial functions, and their ablation results in impaired respiratory chain activity, increased oxidative stress, and mitochondrial DNA damage. Lmx1a/b deficiency caused axonal pathology characterized by α-synuclein+ inclusions, followed by a progressive loss of dopaminergic neurons. These results reveal the key role of these transcription factors beyond the early developmental stages and provide mechanistic links between mitochondrial dysfunctions, α-synuclein aggregation, and the survival of dopaminergic neurons.
Midbrain dopaminergic (mDA) neurons control key functions in the mammalian brain, including voluntary movement, associative learning, and motivated behaviors. Dysfunctions of the dopaminergic (DA) system underlie a wide variety of neurological and psychiatric disorders. The progressive and rather selective degeneration of mDA neurons is one of the principal pathological features of Parkinson’s disease (PD) (1). In PD, neuronal loss is accompanied by the appearance of α-synuclein–enriched intraneuronal inclusions called “Lewy bodies” and “Lewy neurites.” The etiologies of PD remain unsolved, but mitochondrial dysfunction emerges as a central mechanism in inherited, sporadic, and toxin-induced PD (2).
Specification of the subtype identities of mDA neurons begins during embryonic development. The combinatory activation of transcription factors (TFs) and their target genes allows the progenitors to mature progressively and terminally differentiate into postmitotic neuron subtypes. Tremendous efforts have been made to describe the complex spatiotemporal expression of TFs during mDA neuronal development (see refs. 3 and 4 for reviews). After mDA neuron maturation, a large number of developmentally expressed TFs remain active throughout adulthood. Our knowledge of the functional roles of these TFs in mature neurons remains rudimentary. Accumulating evidence shows that transcription factors including the nuclear receptor related 1 protein (Nurr1), En1, Pitx3, Otx2, and Foxa2, which are recognized for their role in the early development of mDA neurons, are also required for the maintenance of phenotypic neuronal identity in the adult (5).
The LIM homeodomain genes Lmx1a/b are early determinants of the fate of mDA progenitors (6), and their actions are essential at each step of DA neuronal generation (7, 8). The murine Lmx1a and Lmx1b proteins are closely related and share an overall amino acid identity of 64%, with 100% identity in their homeodomain and 67% and 83% identity in each LIM domain (9). These neuronal lineage-specific transcription factors control the expression of multiple downstream genes and ultimately determine the morphological, physiological, and functional identity of mDA neurons. It is noteworthy that Lmx1a is part of a minimal transcription factor mixture, along with Mash1 and Nurr1, which is able to generate DA neurons directly from mouse and human fibroblasts without the necessity of reverting to a progenitor-cell stage (10). Lmx1a and Lmx1b continue to be expressed in postmitotic precursors and differentiating mDA neurons, but their functional importance in postnatal life is still unknown. Because human LMX1A/B polymorphism has been associated with PD (11), it is imperative to explore the putative role of Lmx1a and Lmx1b genes in the maintenance of mDA neurons.
In the present study, two different targeting approaches based on the Cre-lox recombination system were used to investigate the function of Lmx1a/b in mature mDA neurons. Our work provides mechanistic insights into the physiological relevance of Lmx1a/b in the adult brain per se and also provides important cues concerning the mechanisms of neuronal degeneration processes. We found that Lmx1a/b are master regulator genes involved in the active maintenance of DA circuits throughout the lifespan. Our results uncover pathways downstream of Lmx1a/b that are involved in regulating the mitochondrial metabolism of mDA neurons. We discuss the relevance of our findings in the context of PD because the disruption of Lmx1a/b regulatory networks in a genetic mouse model recreates some of the cellular features of the disease to an unprecedented level of accuracy.
Results
Expression of Lmx1a and Lmx1b in the Postnatal Brain.
We used in situ hybridization and immunohistochemistry on wild-type mouse midbrain sections to investigate the expression profiles of Lmx1a and Lmx1b throughout the postnatal period (Fig. 1). The Lmx1a hybridization signal was highest at postnatal day 1 (P1) and diminished progressively with aging but still was present at 6 mo (P200) (Fig. 1A). The Lmx1b transcript level was higher at P7 than during the embryonic period and remained expressed at moderate levels throughout all postnatal stages from P1 to P200 (Fig. 1A). Triple-immunofluorescence staining in P200 midbrain sections revealed that nearly all mDA neurons were positive for Lmx1a, Lmx1b, and tyrosine hydroxylase (TH) (Fig. 1B). We confirmed the expression pattern by Western blotting analysis of ventral midbrain extracts. Although the level of expression is reduced in the adult brain as compared with the early postnatal stage, robust protein levels of both Lmx1a and Lmx1b (Fig. 1C and Fig. S1) were still detectable in aged (13-mo-old) mice, supporting previous reports (12–14).
Fig. 1.
Lmx1a and Lmx1b expression in mDA neurons is maintained into adulthood. (A) Lmx1a and Lmx1b transcripts levels were compared in P1, P7, P60, and P200 in C57BL6 mouse ventral midbrain cryosections using nonradioactive in situ hybridization. (Scale bar, 250 μm.) (B) Stitched images of confocal tiles showing immunofluorescent staining with anti-Lmx1a (green), anti-Lmx1b (red), and anti-TH (blue) in ventral midbrain cryosections of P1 and P200 C57BL6 mouse midbrain illustrating the protein expression. (Scale bar, 250 μm.) (C) Western blot and quantification of Lmx1a and Lmx1b expression in E15, P1, P7, P20, adult (3 mo old), and aged (9 mo old) mouse midbrain. n = 3 animals per age. The molecular mass of the protein marker is indicated.
Fig. S1.
(Left) Western blots from ventral midbrain extracts from P7 and 13-mo-old mice. (Right) Quantification of Lmx1a and Lmx1b protein levels (n = 3 animals per age).
Conditional Deletion of Lmx1a and Lmx1b in Mature mDA Neurons Leads to Progressive DA Neuron Loss.
Because Lmx1a and Lmx1b can compensate for the loss of the other (8), we evaluated the involvement of both factors in the maintenance of mature mDA neurons using an Lmx1a and Lmx1b double conditional-knockout (cKO) mutant, Lmx1a/b cKO. To delete both alleles of these genes specifically in mDA neurons without affecting either the specification or the normal proliferation of DA progenitors, we generated a mouse line in which Lmx1a and Lmx1b are conditionally deleted in postmitotic mDA neurons (Fig. S2). We crossed double-homozygous Lmx1a and Lmx1b floxed mice with heterozygous mice expressing Cre recombinase under control of the DA transporter (DAT) gene locus (Dat-Cre). In this line, Cre recombinase is expressed at embryonic day 13.5 (E13.5) in virtually all mDA neurons (15). Littermate DAT+/+ Lmx1a/b F/+ or F/F mice were used as controls, unless noted otherwise in the text.
Fig. S2.
In situ hybridization for Lmx1a and Lmx1b on Lmx1a/b cKO and control midbrain sections at P1 showing the absence of Lmx1a and Lmx1b expression in the Lmx1a/b cKO midbrain. (Scale bar, 500 μm.)
Unbiased stereological and immunohistological analyses of the ventral midbrain were conducted in Lmx1a/b cKO and control mice. Localization, distribution, and neuronal counts of TH+ neurons in the ventral tegmental area (VTA) and the substantia nigra pars compacta (SNpc) showed no observable differences between Lmx1a/b cKO and control mice at P1 (Fig. 2 A, D, and E). At P20, stereological neuronal counts showed no differences in the number of TH+ neurons in the VTA (Fig. 2 B and D), but we measured a significant neuronal loss in the SNpc of Lmx1a/b cKO mice (Fig. 2 B and E). In 2-mo-old (P60) Lmx1a/b-deficient mice, we measured a 35% reduction in the number of TH+ neurons in the VTA and a 44% reduction in the SNpc (Fig. 2 D and E). Accordingly, the density of DA dendrites that normally penetrate deep into the substantia nigra pars reticulata was greatly reduced in Lmx1a/b cKO mice (Fig. 2C). The number of TH+ neurons in the VTA and SNpc did not decrease substantially between P60 and P200 (around 6.5 mo), because the percentage of reduction in both the VTA and SNpc remained similar in Lmx1a/b cKO and control mice.
Fig. 2.
Effects of Lmx1a and Lmx1b ablation on mDA neurons. (A–C) Immunohistological analyses of TH staining in the VTA and SNpc in the midbrains of P1 (A), P20 (B), and >P60 (C) Lmx1a/b cKO and control mice using DAB staining against TH. The arrows in C denote the DA neuronal loss. (Scale bar, 500 μm.) (D and E) Quantification of DA neuronal loss in the VTA (D) and SNpc (E) by stereological count at P1, P20, P60, and P200; values are derived from at least four mice, three sections per brain. (F) Coexpression of DAT (green) and TH (red) in midbrain sections from 60-d-old Lmx1a/b cKO and control mice (stitched confocal images). Arrows indicate that the loss of TH+ cells is correlated with a loss of the DAT signal. (Scale bar, 500 μm.) (G) β-Gal staining (blue) on midbrain sections from 60-d-old Lmx1a/b cKO R26RLacZ/+ and control mice confirming the loss of DA neurons. The genotype of control mice in G is DatCre/+R26RLacZ/+. (Scale bar, 500 μm.)
To confirm the complete loss of TH neurons, stereological analysis for DAT- and Nissl-stained cells was conducted at P60. The loss of TH+ neurons correlated with the loss of the DAT and Nissl signal (Fig. 2F and Fig. S3). We also used a LacZ reporter line to visualize neuronal loss. We crossed Lmx1a/b cKO mice with a Rosa26 LacZ reporter line (16). In these mice, LacZ is conditionally expressed in mDA neurons that have undergone Cre-mediated excision. The ventral midbrain of the Dat-Cre control mice was densely packed with cells expressing β-gal. By contrast, only sparse β-gal+ neurons were scattered within the VTA and SNpc of Lmx1a/b cKO mice, confirming the loss of mDA neurons (Fig. 2G).
Fig. S3.
(A and B) Immunohistological analyses of DAT and Nissl staining in coronal midbrain sections from control (A) and Lmx1a/b cKO (B) mice using DAB staining against DAT. (Scale bar, 500 μm.) (C and D) Higher magnification images of SNpc sections from control (C) and Lmx1a/b cKO (D) mice. (Scale bar, 250 μm.) (E) Quantification of DA neuronal loss in the VTA and the SNpc by unbiased stereological counting of TH+, DAT+, and Nissl+ cells at P60. For the DAT and Nissl quantification, we selected the region of interest (mDA domain) based on DAT staining and counted the DAT+- and Nissl+-stained cells in this region. Values are derived from at least four mice, three sections per brain.
Abnormal Accumulation of α-Synuclein in mDA Nerve Terminals of Lmx1a/b cKO Mutant Mice.
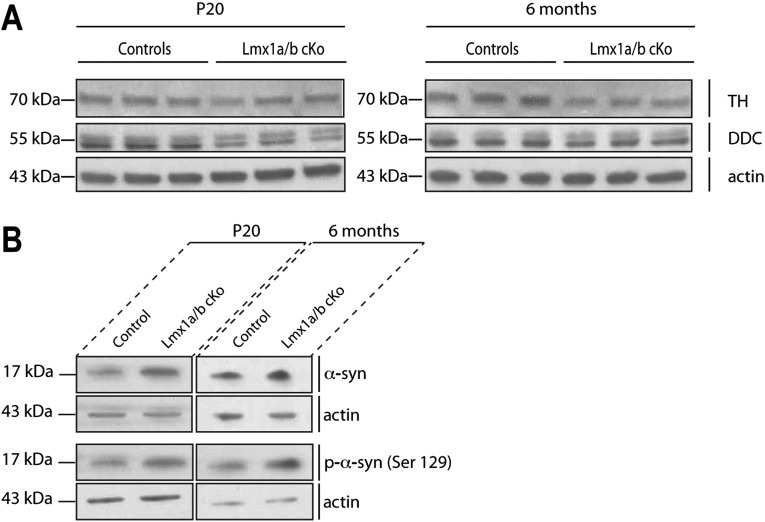
In accordance with the difference in the number of TH+ cell bodies in the SNpc at P20 (Fig. 2E), a reduction in TH immunoreactivity was observable in the striatum of Lmx1a/b cKO mutant mice (Fig. 3 A and B). This result was further confirmed by Western blotting analysis of striatal homogenates which revealed an ∼50% decrease of TH and DOPA decarboxylase (DDC) protein levels (Fig. 3E and Fig. S4) at P20. The TH+ nerve fibers in the striatum of mutant mice appeared dystrophic, and abnormal axon terminal enlargements were observed (Fig. S5 A and B). Dystrophic neurites were present as early as P20, and fiber pathology was aggravated with aging. The most striking finding in the striatum of Lmx1a/b cKO mice was the abundance of large TH+ spheroid-like structures (Fig. 3B). Immunohistological staining from P20 and 2-mo-old mice showed numerous TH+ puncta of various sizes (5–20 μm), accumulating predominantly in dystrophic nerve fibers in the striatum. Double immunostaining experiments with antibodies directed against aggregation-prone proteins were performed to identify the composition of the puncta. Puncta were found to be α-synuclein immunoreactive (Fig. 3 C and D) at P60. α-Synuclein is a small presynaptic protein well known for its close association with PD pathology. Lewy bodies and Lewy neurites observed in PD are largely composed of abnormal fibrillar aggregates enriched with a phosphorylated form of α-synuclein. Striatal tissue extracts were assayed by Western blotting to determine whether phosphorylated α-synuclein (phospho-Ser129 α-synuclein) was present. The ratio of phospho-Ser129 α-synuclein to total α-synuclein from Lmx1a/b cKO mice aged 6 mo or older was increased significantly compared with controls (Fig. 3F and Fig. S4). In accordance with the abnormal DA nerve terminals found in the striatum, HPLC of striatal extracts confirmed the reduction of DA and DA metabolites in mutant animals (Fig. 3 G and H). Scattered TH+ cell bodies were also found in the striatum of Lmx1a/b cKO mice, a compensatory mechanism previously reported in the striatum from PD brains (Fig. S5C) (17, 18).
Fig. 3.
Abnormal nerve terminals in Lmx1a/b cKO mice. (A and B) The immunohistological striatal TH staining pattern was compared in P60 control (A) and Lmx1a/b cKO (B) mice. In B, the higher-magnification image shows dystrophic and abnormal axon terminal enlargements of TH+ nerve fibers as well as large TH+ spheroid-like structures. (Scale bar, 50 μm.) See also Fig. S5. (C and D) Identification of the axonal spheroids using coimmunostaining of α-synuclein (green) and TH (red) in the striatum of control (C) and Lmx1a/b cKO (D) mice. (Scale bar, 50 μm.) (E and F) Western blotting analysis of TH, DDC, and phospho-α-synuclein levels in extracts from the striatum of P20 (E) and 6-mo-old (F) control and Lmx1a/b cKO mice (n = 3 animals per genotype). Representative blots are shown in Fig. S4. (G and H) HPLC analysis of striatal content (DA, DOPAC, 3-MT, and HVA) in P20 (G) and 6-mo-old (H) control and Lmx1a/b cKO mice. Values are derived from at least three mice per group. (I) Lmx1a/b cKO mice and their controls were placed in the open-field chamber. Horizontal and vertical activities (measured by beam breaks) and the total distance traveled were measured for 10 min in P30, P60, P90, P200, and P300 mice.
Fig. S4.
(A) Western blotting analysis of TH and DDC levels in extracts from the striatum of control and Lmx1a/b cKO mice at P20 and at age 6 mo (n = 3 animals per genotype). The molecular mass of the protein marker is indicated. (B) Western blotting analysis of α-synuclein and phospho-α-synuclein S129 levels in extracts from the striatum of control and Lmx1a/b cKO mice at P20 and at age 6 mo (n = 3 animals per genotype). The molecular mass of the protein marker is indicated.
Fig. S5.
(A) Higher-magnification images of the striatum from P20 Lmx1a/b cKO and control mice show representative TH+ spheroid-like structures present in Fig. 3 A and B. (Scale bar, 50 μm.) (B) Quantification of the number of axonal spheroids in the striatum from P20 and P60 Lmx1a/b cKO and control mice; values are derived from at least three mice with two striatal complete sections manually counted per brain (***P ≤ 0.001). (C) Examples of sporadic TH+ cell bodies (arrows) found in the striatum of Lmx1a/b cKO mice at P60. TH (green) was colabeled with nuclear staining DAPI (blue). (Scale bar, 50 μm.)
Next, we analyzed the spontaneous locomotor activity of the Lmx1a/b cKO mutant mice in an open-field chamber at different ages. The Lmx1a/b cKO mice were hyperactive. The total distances traveled were significantly affected in mutant mice and also gave the clearest signal of a progressive decline in locomotor activity with aging (Fig. 3I).
Degeneration of mDA Neurons Following Adeno-Associated Virus–Mediated Cre Inactivation of Lmx1a/b in Adult Mice.
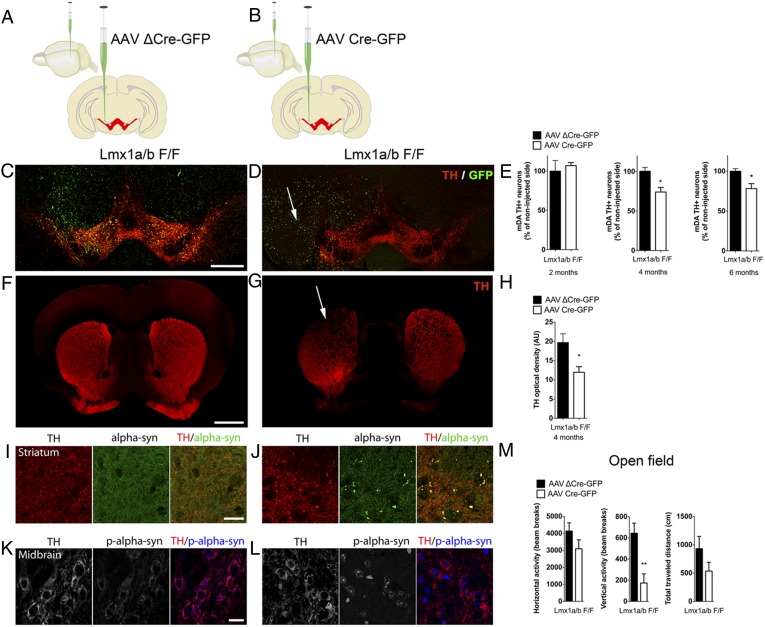
To eliminate putative confounding effects of compensatory postnatal developmental mechanisms, we deleted Lmx1a/b in mDA neurons using viral delivery of Cre recombinase through stereotaxic injection in the midbrain of adult mice (Fig. 4 A and B and Fig. S6). Mice that received unilateral intra-SNpc/VTA injections of a Cre virus recapitulated the essential neuropathological features observed in Lmx1a/b cKO mice (Fig. 4 C, D, F, and G). Stereological analysis of TH+ cells showed no difference in neuronal count 2 mo after injection. However, a significant decrease was measured 4 mo and 6 mo after the injection of adeno-associated virus (AAV) Cre (Fig. 4E). Four months following stereotaxic infusion of the Cre virus, TH optical density was reduced throughout the striatum on the lesioned side of AAV Cre- injected animals (Fig. 4 F–H). Dystrophic swollen TH+ neurites and large spheroid-like structures were also observed in the striatum on the lesioned side of the brain (Fig. 4 I and J). TH and α-synuclein colocalized in these spheroids, in contrast to the diffused distribution seen in the striatum of the animals injected with control virus (Fig. 4 I and J). To verify the progression of the process, we also stained TH and α-synuclein at an earlier time point (2 mo postinjection); at this stage, TH and α-synuclein do not colocalize in these spheroids (Fig. S7A). α-Synuclein undergoes several posttranslational modifications. For example, phosphorylation at serine 129 (pS129) has frequently been linked to PD pathogenesis because it may increase the formation of α-synuclein aggregates. At the midbrain level, a difference in pS129 content and subcellular localization was observed between control and AAV Cre-injected animals (Fig. 4 K and L). Additionally, AAV Cre-injected animals demonstrated a significant decrease in vertical activity (a decrease of nearly 75% compared with control mice) 4 mo following the virus injection (Fig. 4M). To evaluate the influence of the age of ablation on the degenerative process, we injected AAV Cre vector in young (20-d-old) and aged (20-mo-old) mice. Locomotor measurements indicate that young mice cope better with the loss of Lmx1a/b, because no motor dysfunction was observed in these mice 3 mo after Cre delivery. By contrast, aged mice showed significant motor impairment in horizontal activity, vertical activity, and total distance traveled only 1 mo after the AAV Cre injection (Fig. S8).
Fig. 4.
Inactivation of Lmx1a and Lmx1b in mDA neurons by viral-vector delivery of Cre recombinase impairs the survival of adult midbrain DA neurons. (A and B) Schematics depicting the injection of control mutated Cre-encoding (AAV ΔCre-GFP) virus (A) or of Cre-encoding (AAV Cre-GFP) virus (B) in the ventral midbrain of Lmx1a/b F/F adult (2-mo-old) mice. (C and D) Stitched confocal images of TH and GFP staining from a representative section of Lmx1a/b F/F mice 6 mo after unilateral injection with either control or Cre-encoding (AAV Cre-GFP) virus. Infected cells are shown in green, and the arrow denotes the DA neuronal loss. (Scale bar, 500 μm.) (E) Quantification of DA neuronal loss in the VTA and the SNpc by stereological counting of TH+ cells at 2, 4, and 6 mo after the injection; values are derived from at least four mice, three sections per brain. (F and G) TH staining in the striatum (red) shows the DA axon distribution (stitched confocal images). The arrow indicates the DA axonal loss in the dorsolateral striatum on the injected side. (Scale bar, 1 mm.) (H) Densitometry quantification of TH immunostaining intensity in the striatum of mice infected with the control (mutated Cre) and Cre-GFP vectors (n = 3). (I and J) Identification of the axonal spheroids using costaining of α-synuclein (green) and TH (red) in the striatum of the side injected with either control or Cre-encoding AAV. (Scale bar, 50 μm.) (K and L) Immunohistological analysis of phosphorylated α-synuclein at S129 (blue) in TH+ neurons (red) in the midbrain of injected mice. High levels of phospho α-synuclein S129 were detected in the nucleus of TH+ neurons in AAV Cre-GFP–injected animals compared with controls. (M) Behavioral characterization of Lmx1a/b F/F mice 6 mo after AAV infection in the SNpc. Open-field horizontal and vertical activity (measured by beam breaks) and total distance traveled were measured (n = 5).
Fig. S6.
(A and B) In situ hybridization of Lmx1a and Lmx1b (A) and β-gal (B) staining on midbrain sections 2 mo following injection of AAV ΔCre-GFP, AAV Cre-GFP, and AAV Cre-GFP-ires-Nrf1 (stitched images). The absence of Lmx1a and Lmx1b staining at the injection site shows the efficiency of the AAV vector used. (C) In vitro validation of AAV Cre-ires-Nrf1 in HEK293 cells. The HEK293 cell line was infected, mRNA was extracted 2 wk later, and the Nrf1 transcript level was measured by RT-qPCR. (D) RT-qPCR analysis showing Nrf1 mRNA expression in primary neuronal cultures of ventral midbrain from P1 control or Lmx1a/b cKO mice transfected with a control empty vector or a vector expressing Nrf1 (CMV-Nrf1) (n = 3). (E) Expression profile of Lmx1a and Lmx1b using reverse-transcription PCR. Lmx1a and Lmx1b amplification products (349 pb and 307 pb, respectively) obtained from noninjected and injected (asterisks) hemispheres of dissected ventral midbrain showing a lower amount of Lmx1a and Lmx1b transcripts in the hemispheres injected with the AAV Cre-GFP (2) and AAV Cre-GFP-ires-Nrf1 (3). Note that levels of Lmx1a and Lmx1b were similar in both hemispheres following AAV ΔCre-GFP (1) injection.
Fig. S7.
(A) Identification of the axonal spheroids using costaining of α-synuclein (blue) and TH (red) in the striatum of the midbrain side injected with either control or Cre-encoding AAV, 2 mo postinjection. Note that axonal spheroids do not show α-synuclein accumulation 2 mo postinjection. (Scale bar, 50 μm.) (B) Dystrophic and abnormal axon terminal enlargements of TH+ nerve fibers as well as large TH+ spheroid-like structures in mice injected with AAV Cre. No abnormal axon terminal enlargements were found in mice injected with AAV ∆Cre and AAV Cre-Ires-Nrf1 up to 6 mo after injection. (Scale bar, 50 μm.)
Fig. S8.
(A and B) Mice that were injected with AAV ∆Cre and AAV Cre at P20 were placed in the open-field chamber. Horizontal and vertical activities (measured by beam breaks) and the total distance traveled were measured for 60 min at 1 mo (A) and 3 mo (B) postinjection. (C and D) Mice that were injected with AAV ∆Cre and AAV Cre at age 20 mo were placed in the open-field chamber. Horizontal and vertical activities (measured by beam breaks) and the total distance traveled were measured for 60 min at 1 mo (C) and 3 mo (D) postinjection.
Lmx1a/b Coordinate the Expression of Mitochondrial-Associated Genes.
In an attempt to elucidate the mechanisms by which Lmx1a and Lmx1b participate in the postnatal maintenance of mDA neurons, we next investigated potential relevant target genes. We first used laser-capture microdissection (LCM) to isolate TH-stained neurons of the SNpc and VTA from Lmx1a/b mutant and control mice at E15.5 followed by next-generation RNA-sequencing (RNA-seq) to obtain the differentially expressed genes. We found 517 genes differently expressed by at least twofold between mutant and control mice. Gene Ontology (GO) analysis was performed to identify possible enrichment genes with specific biological themes. A spectrum of biological processes was significantly enriched, including the mitotic cell-cycle checkpoint, regulation of the cell cycle, and protein–DNA complex assembly, as is consistent with the established roles for developmental transcription factors (Fig. 5B). GO classification also showed that overrepresentation of genes associated with mitochondrial processes such as the long-chain fatty acid metabolic process, acyl-CoA metabolic process, electron transport chain, protein folding, and organelle organization was repressed in mutant mice (Fig. 5B). By comparing the differentially expressed gene list with microarray analysis of LMX1A-overexpressing MN9D DA cells (19), we identified several putatively regulated genes known to be relevant in mitochondrial functions (Table S1). We also searched our RNA-seq list to find genes related to previously identified molecular interactors of LMX1B (20) and found candidate genes involved in stress protection. Next, we validated each candidate gene as an Lmx1a/b target by RT quantitative PCR (RT-qPCR) from laser-microdissected mDA neurons from P1 control and Lmx1a/b cKO mice (Fig. 5A) (21).
Fig. 5.
Lmx1a/b coordinate the expression of mitochondrial-associated genes. (A) Schema illustrating rapid TH immunostaining, LCM, and mRNA isolation. (Scale bars, 500 μm.) (B) Enrichment of GO terms for biological processes associated with down-regulated genes in Lmx1a/b cKO. (C) RT-qPCR analysis showing Nrf1, Ndufa2, Ndufa3, Ndufa4, Ndufv1, Uqcrq, Cox1, Cox6a, and Hspa8 mRNA expression in the ventral midbrain of control and Lmx1a/b cKO mice at P1. (D) RT-qPCR analysis showing Lmx1a, Lmx1b, and Nrf1 mRNA expression in mDA cultured neurons overexpressing Lmx1a (n = 5) and Lmx1b (n = 7). (E) Respirometry experiments in mDA cultured neurons from controls and Lmx1a/b cKO mice. CCCP, oxygen consumption rate in the presence of CCCP; OCR, oxygen consumption rate; RCR, respiratory control ratio. Mean basal OCR was 600 and 444 pM⋅min−1⋅104 neurons−1 for control and Lmx1a/cKO neurons, respectively, and the mean for uncoupled OCR was 976 and 601 pM⋅min−1⋅104 neurons−1 for control and Lmx1a/cKO neurons, respectively. The graph at the right compares the survival of DA neurons from control and Lmx1a/b cKO mice after 10 DIV (n = 5).
Table S1.
Differential expression of genes of interest identified by RNA sequencing and RT-qPCR
| Gene | RNA-seq at E15.5, fold change | RT-qPCR at P1, fold change |
| Nrf1 | −1.45 | −2.36 |
| Ndufv1* | −2.27 | −1.29 |
| Ndufa3* | −3.00 | −1.87 |
| Ndufa2 | −1.49 | −8.06 |
| Ndufa4 | 1.19 | −9.17 |
| Uqcr (Uqcr11) | 2.70 | −1.13 |
| Uqcrq* | −2.23 | −1.75 |
| Cox1 | No data | −1.02 |
| Cox6a1 | −1.98 | −2.75 |
| Mrps35 | −1.72 | −1.46 |
| Mrps31 | −1.68 | −1.31 |
| Hspa8* | −2.54 | −1.51 |
Identified in the GO analysis (Fig. 5B).
mRNA levels of the mitochondrial complex I genes Ndufa3, Ndufa2, and Ndufa4, the mitochondrial complex III gene Uqcrq, and the mitochondrial complex IV gene Cox6a, all of which are nuclear-encoded subunits of the respiratory chain, were significantly lower in LCM-dissected tissue from Lmx1a/b cKO mice (Fig. 5B). Although it has not been identified as a target of Lmx1a/b, we included the mitochondrial Cox1 gene in our analysis as a mitochondrial DNA marker to verify whether the difference in the mRNA level observed in the respiratory chain subunits was associated with a change in mtDNA and/or mitochondrial quantity. No significant difference in the Cox1 gene level was detected between control and Lmx1a/b cKO mice, suggesting the absence of a large change in mitochondrial number. Nuclear respiratory factor-1 (Nrf1), an essential transcription factor for the integration of nuclear- and mitochondrial-encoded gene transcription (22), was differentially expressed in control and mutant mDA tissue, with a highly significant decrease in Lmx1a/b cKO mice. These observations suggest that Lmx1a/b control the expression of both mitochondrial transcription factor (Nrf1) and nuclear-encoded mitochondrial genes. To confirm the correlation between Lmx1a/b deficiency and Nrf1 down-regulation, we forced the expression of Lmx1a and Lmx1b in primary neuronal cultures through transient transfection. Three days after overexpression of Lmx1a, a threefold increase in Nrf1 expression was detected (Fig. 5C). Overexpression of Lmx1b also resulted in a reproducible and robust 15-fold up-regulation of Nrf1 (Fig. 5D).
We also investigated the Hspa8 gene, which encodes the heat shock cognate 70 (Hsc70) protein, the constitutively expressed form of Hsp70. Immunoprecipitation experiments identified Hsp70 as a likely interactor with Lmx1b (20). Hsc70 is a specific chaperone known to interact with the PD-associated protein α-synuclein (23, 24). A significantly lower mRNA level of Hspa8 was measured in Lmx1a/b cKO mice (Fig. 5C).
Deficiency in Lmx1a/b Leads to Impaired Mitochondrial Respiration and DA Neuronal Degeneration in Vitro.
To determine whether the alteration in transcriptional regulation by Lmx1a/b had any functional consequence on mitochondrial bioenergetics, we next performed respirometry experiments on 10-d-old mDA neurons cultured from control and Lmx1a/b cKO mice. We assessed the respiration rate as measured by basal and maximal [uncoupled with carbonylcyanide-m-chlorophenylhydrazone (CCCP)] oxygen consumption rate (OCR, measured in picomoles of oxygen per minute) using a Seahorse extracellular flux analyzer. Cellular respiration was measured in live neurons to estimate mitochondrial oxidative phosphorylation. These experiments demonstrated that the basal OCR was significantly decreased in Lmx1a/b cKO neurons, as is consistent with the decreased mitochondrial respiration in these cells (Fig. 5E). This decrease in respiration could be attributable to a lower mitochondrial density or to a reduced oxidative capacity of mitochondria. Furthermore, the maximal respiratory capacity was significantly lower in neurons from Lmx1a/b cKO mice than in neurons from controls. The respiratory control ratio, i.e., the ratio of maximal to basal OCR, was significantly lower in neurons from Lmx1a/b cKO mice than in neurons from controls, indicating a lower spare mitochondrial capacity. These data suggest the involvement of a mitochondrial dysfunction at the respiratory chain level rather than a reduction in mitochondrial density. Together, these results indicate that in Lmx1a/b cKO mice the mDA neurons have less potential respiratory capacity to face increased energetic demands and/or various stresses. Likewise, the viability of Lmx1a/b-deficient neurons in primary DA neuronal cultures also revealed their higher vulnerability. After 10 d, a 25% reduction in mDA neuronal survival was measured in primary DA neuron cultures from Lmx1a/b cKO mice compared with controls (Fig. 5E).
Lmx1a/b Mutant Cells Exhibit Overproduction of Reactive Oxygen Species.
Defective function at the level of the mitochondrial electron transport chain could lead to excessive production of reactive oxygen species (ROS). Given the observed mitochondrial impairment in midbrain neurons of Lmx1a/b cKO mice, we next used primary neuronal cultures of the ventral midbrain from controls and Lmx1a/b cKO mice to examine ROS production. Dissociated P1 midbrain was cultured at low density, a condition under which mDA neurons undergo gradual cell death. A live-cell imaging technique using a redox-sensitive probe (CellROX) after 3 d of culture revealed a significantly higher rate of ROS production in Lmx1a/b cKO cells than in controls (Fig. 6 A–C and Fig. S9A). These results suggest that the Lmx1a/b mutant cells could generate increased ROS levels under stress conditions.
Fig. 6.
Deficiency in Lmx1a/b leads to overproduction of ROS and impaired mitochondrial DNA, and forced expression of Nrf1 has a rescue effect both in vitro and in vivo. (A) Primary neuronal cultures of ventral midbrain from P1 control or Lmx1a/b cKO mice transfected with either control empty vector or a vector expressing Nrf1 (CMV-Nrf1). Redox-sensitive probes (CellROX; green) were used. (Scale bar, 200 μm.) (B and C) Densitometry quantification [corrected total cell fluorescence (CTCF)] of the ROS-sensitive probe (B) and relative percentage of TH+ neurons surviving (C) after 3 DIV in Lmx1a/b F/F primary neuronal cultures where control (CMV-empty) and a Nrf1-expressing vector (CMV-Nrf1) were transfected (n = 5). (D) Representative images (stitched confocal images) of the SNpc from P60 control and Lmx1a/b cKO mice. Tissue was stained for TH (red) and a mitochondrial marker Tom20 (blue) and was probed with an ARP (green) (n = 3). (Scale bar, 20 μm.) (E) TH and GFP labeling from a representative section of Lmx1a/b F/F mice 2 mo after unilateral injection with either AAV Cre-GFP or AAV Cre-GFP-Ires-Nrf1 (stitched confocal images). (Scale bar, 500 μm.) (F) Higher-magnification images of the midbrain from injected mice show no obvious abnormalities in the cell morphology and TH+ neuron distribution following injection of Cre-Nrf1 vector, whereas abnormal and dystrophic neurons were discernible in Cre-injected mice 2 mo after injection. (Scale bar, 20 μm.) (G) Stereological neuronal counting of TH immunoreactive neurons. Data are presented as the percent of mDA neurons in the injected side relative to the noninjected side 6 mo after injection (n = 4). (H) DA axons appear normal in the striatum of mice injected with AAV Cre-GFP-ires-Nrf1. There is no sign of axonal spheroids enriched in α-synuclein 2 mo after injection. (Scale bar, 100 μm.) (I) Immunohistological labeling of phosphorylated α-synuclein at S129 (blue) and TH+ neurons (red) in the midbrain of injected mice. (Scale bar, 20 μm.) (J) Behavioral measurements of horizontal and vertical movements and total traveled distance using an open-field chamber 6 mo after injection.
Fig. S9.
(A) Densitometry quantification (CTCF) of a ROS-sensitive probe in TH+ neurons. Primary neuronal cultures of ventral midbrain from P1 control or Lmx1a/b cKO mice were cotransfected with CMV-empty and CMV-mCherry vectors as negative control or with an Nrf1-expressing vector (CMV-Nrf1) and CMV-mCherry. Cultured neurons were treated with redox-sensitive probe (CellROX) (green) and were fixed and immunostained against TH. The probe intensity values were measured in individual neurons that were transfected (mCherry) and were TH+ (n = 14). (B) Transfection efficiency and TH+ neuron number in primary culture. Dissociated primary neurons from the midbrains of Lmx1a/bF/F mice were transfected at DIV 2 with CMV-mCherry. One day following transfection, cells were fixed and stained for TH and Map2. Fifty-four percent of the neurons (Map2) were transfected, and 44.8% of the neurons were TH+.
Deficiency in Lmx1a/b Leads to mtDNA Damage.
Oxidative damage to mitochondrial DNA is observed in several age-associated disorders, including PD. Given the mitochondrial impairment and increased oxidative stress observed in midbrain neurons of Lmx1a/b cKO mice, we next evaluated damage to mtDNA. Abasic sites are a type of DNA damage, often arising from oxidative stress, in which loss of a purine or pyrimidine base occurs (25). Using an adapted histochemical assay (25), elevated levels of abasic sites were measured in mDA neurons of Lmx1a/b cKO mice (Fig. 6D). In mature (2-mo-old) Lmx1a/b cKO mice, more than 64% of DA neurons demonstrated a striking ARP (aldehyde-reactive probe) fluorescence signal. In control mice, the ARP signal was fainter or absent in the majority of DA neurons. The colocalization of the ARP signal with the mitochondrial marker Tom20 demonstrated that accumulated abasic sites were specific to mitochondrial DNA and not to nuclear DNA. Accelerated accumulation of somatic mtDNA mutations in mDA neurons could contribute to the neurodegeneration observed in Lmx1a/b cKO mice.
Altered Autophagy in Lmx1a/b cKO Mice.
Evidence suggests the involvement of autophagy deregulation in PD, and a recent study reported that removal of Lmx1b in postnatal mDA neurons produced enlarged axon terminals associated with an alteration in autophagy (13). In 2-mo-old Lmx1a/b cKO mice, we measured and confirmed a significant change in LC3-II at the protein level (Fig. S10 A and B). A decrease in the LC3-II signal may indicate either a decrease in autophagosomal synthesis or an increase in autophagosomal clearance. In either case, our results suggest a modified autophagic flux. However, according to the RNA sequencing data at E15.5 and qPCR results at P1, genes of the autophagy–lysosomal pathway do not appear to be modified in Lmx1a/b cKO mice (Fig. S10).
Fig. S10.
(A) Western blotting analysis of LC3-I and LC3-II levels in extracts from the striatum of control and Lmx1a/b cKO mice at P60 (n = 3 animals per genotype). The molecular mass of the protein marker is indicated. (B) Quantification of the relative density of LC3-II. (C) E15.5 mRNA sequencing analysis for the autophagic–lysosomal pathway. Values are shown in RPKM. (D) RT-qPCR analysis showing Atg5, Becn1, and Lamp1 mRNA quantification from LCM mDA neurons of control and Lmx1a/b cKO mice at P1 (n = 3 animals per genotype).
Forced Expression of Nrf1 Rescues Aberrant ROS Levels in Vitro and both Cellular and Locomotor Deficits in Vivo.
To establish a relationship between the absence of Lmx1a/b and mitochondrial dysfunction, we chose to restore (and increase) the expression of Nrf1. Through the transcriptional control of several mtDNA genes and nuclear genes (putatively including NDUFA2, NDUFA3, and NDUFA4), Nrf1 holds a dominant position in the hierarchy of mitochondrial homeostasis. We first restored Nrf1 expression in primary cultures from Lmx1a/b mutant and control cells through transient transfection. Forced expression of Nrf1 in DA neuronal cultures rescued abnormal production of ROS in Lmx1a/b mutant cells (Fig. 6 A and B). Furthermore, Nrf1 overexpression significantly increased the overall survival of mDA neurons from both mutants and controls (Fig. 6C). These in vitro findings suggest that Nrf1 overexpression might reduce ROS production. To investigate the role of Nrf1 in the regulation of mDA neuron maintenance further, rescue experiments were performed in which Cre-mediated inactivation of Lmx1a/b was combined with the forced expression of Nrf1. We generated a biscistronic vector coexpressing the functional Cre and Nrf1 (AAV Cre-GFP-Ires-Nrf1) and injected it unilaterally into the ventral midbrain of Lmx1a/b double-floxed mice (Fig. 6E and Fig. S6). No obvious abnormalities were detected in the cell morphology and TH+ neuron distribution 2 mo following injection of the Cre-Nrf1 vector, but abnormal and dystrophic neurons were already discernible in Cre-injected mice (Fig. 6F and Fig. S7). Histological analysis confirmed that the forced expression of Nrf1 rescued mDA neuronal loss (Fig. 6G). Neither dystrophic swollen TH+ neurites nor large spheroid-like structures were observed in the striatum on the lesioned side of the brain (Fig. 6H). Moreover, 6 mo postinjection, the striatum of mice injected with AAV Cre-GFP-Ires-Nrf1 showed only a few abnormal terminals and no obvious signs of puncta (Fig. S7B). Immunohistological analysis of phosphorylated α-synuclein in TH+ neurons in the midbrain revealed no obvious pattern of accumulation or aggregation (Fig. 6I) as seen following AAV Cre injection (Fig. 4L). Locomotor abnormalities caused by Lmx1a/b deficiencies were fully rescued by the forced expression of Nrf1 6 mo following injection (Fig. 6G and Fig. S7).
Discussion
Here we show that maintenance of mDA neurons in adulthood is severely compromised in the absence of Lmx1a/b. This finding indicates critical roles for Lmx1a/b in the adult brain and also offers unique insights into active maintenance mechanisms and DA neuron degeneration.
Our results describe the functional consequences of Lmx1a/b ablation in mature mDA neurons. In the Lmx1a/b cKO (Dat-Cre) mice, signs of degeneration first appear at P20 in the SNpc concomitant with the appearance of abnormal swollen terminals (called “spheroid-like structures”) preceding mDA neuronal death. PD is characterized by the degeneration of the mDA neurons, largely those of the SNpc. We observed a progressive loss of mDA neurons and a depletion of DA in the striatum of these mice from the age of 3 wk to 2 mo. The axonal spheroid-like structures that consistently characterized neurons lacking Lmx1a/b in these mice around age 2 mo were found to be immunoreactive for α-synuclein. A prominent pathological hallmark of PD is the presence of Lewy bodies, which are intraneuronal inclusions principally composed of fibrillized α-synuclein (26). After 2 mo and up to 6 mo, no further substantial neuronal loss was observed. The locomotor activity of the Lmx1a/b cKO (Dat-Cre) mouse model is perturbed but is difficult to interpret because the mice are hyperactive. However, the total distance they traveled declined constantly with aging in measurements taken up to age 1 y. Because Lmx1a/b regulate several genes during development of postmitotic mDA neurons, it is possible that the hyperactive phenotype could come from a developmental defect. According to our mRNA sequencing data, Slitrk family members seem to be regulated by Lmx1a/b. Although the function of these genes in mDA neurons remains unknown, these proteins have been shown to affect neurite outgrowth (27) and are involved in the formation of regulation synapses (28). Human genetic studies also have linked rare mutations in SLITRK genes with obsessive–compulsive spectrum disorders and other neuropsychiatric disorders including attention deficit hyperactivity disorder (29). A change in the expression of genes important for the correct wiring or for the synaptogenesis of mDA neurons could indeed have diverse consequences on behavior (including hyperactivity), but further studies would be required to confirm this hypothesis. To help clarify the action of Lmx1a/b in the adult brain, we used unilateral injections of AAV vector to deliver Cre recombinase to adult mDA neurons. The progression and main features of the degenerative process observed in these mice are consistent with the Dat-Cre mice model but without any hyperactivity. However, observations of this model suggest that the appearance of spheroid-like structures in the striatum precedes mDA neuronal death. Axonal degeneration is an early feature of PD supported by postmortem studies, functional neuroimaging, and toxin-induced animal models (30). The sequence of pathological events we observed in Lmx1a/b cKO mice is consistent with the retrograde degeneration model. These data suggest that, as in the dying-back PD hypothesis (31), nigrostriatal degeneration is revealed first by perturbation of axon terminals of the striatum. Inactivation of Lmx1a and Lmx1b in mDA neurons by viral-vector delivery also allowed the observation of a progressive, PD-like deterioration of motor function when Lmx1a/b were ablated at age 2 mo or 2 y. We first observed a reduction in vertical movements at 12 wk, and mice showed a continuous decline over the following months. Interestingly, the progression of the observed motor symptoms (i.e., vertical movements declining earlier and faster than the horizontal movements) followed a pattern similar to that reported in the MitoPark PD mice model (32, 33). This behavioral phenotype has been suggested to model the early occurrence of axial/postural instability in PD (32). According to the locomotor measurements, the age of Lmx1a/b ablation seems to be critical. We observed a drastic locomotor deficit when Lmx1a/b were inactivated in aged mice. By contrast, we could not detect significant changes in locomotor behavior in young mice 3 mo after Lmx1a/b inactivation.
Collectively, our results provide compelling evidence that Lmx1a/b cKO mice could serve as a model of PD, combining two major pathological features of the disease, i.e., Lewy neurite-like α-synuclein accumulations and significant mDA neuron degeneration.
Dismantling the action of transcription factors that promote or repress the activity of thousands of genes is likely to have complex consequences, and the functional importance of Lmx1a/b is yet to be fully understood. Here, we provide evidence suggesting that Lmx1a/b can modulate mitochondrial metabolism. We propose that Lmx1a/b control key parts of the program that drives the generation and maintenance of DA neurons. This program includes not only the essential components belonging to the DA pathway but also the unique mitochondrial features of mDA neurons and most likely the constitutive protection required for their survival (34). Therefore, Lmx1a/b might be a key player in the cross-talk between genetic and metabolic signaling that occurs during development, a capacity that persists in mature mDA neurons.
Among the mitochondrial impairments contributing to the etiology of PD, complex 1 deficit seems to be the principal contributor (35). Mitochondrial complex I (NADH ubiquinone oxidoreductase) is the first and largest complex of the respiratory chain. Key genes coding for mitochondrial complex I (i.e., Ndufa2, Ndufa3, and Ndufa4) are down-regulated in Lmx1a/b cKO mice. NDUFA3 has recently been found necessary for the formation of a functional holoenzyme (36). NDUFA2 and NDUFA4 mutations have been linked to Leigh disease, a severe neurological disorder. NDUFA2, NDUFA3, NDUFA4, UQCRQ, and COX6A have all been identified in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map representing the causative gene and molecular network in PD (www.genome.jp/kegg/).
The observation that key genes coding for electron transport complex I and complex III proteins are specifically down-regulated in Lmx1a/b cKO mice supports the hypothesis of a link between these TFs and mitochondrial metabolism. We also found a down-regulation of Nrf1 in Lmx1a/b cKO mice. Nrf1 occupies an upstream position in the hierarchy of transcription factors coordinating mitochondrial biogenesis and function. Nrf1, in collaboration with PGC-1α, controls oxidative phosphorylation through the expression of many key nuclear-encoded mitochondrial genes, including all cytochrome c oxidase nuclear-encoded subunit genes (37, 38). Interestingly, several PD-related genes such as PARK2 (Parkin), PARK6 (Pink1), PARK7 (DJ-1), and PAELR (GPR37) have been identified as NRF1 targets (39). These data indicate that Lmx1a/b activity could influence mitochondrial functions, either directly or indirectly through Nrf1 (or other transcriptional cascades). Consistent with these observations, our oxygen consumption measurements in neurons from Lmx1a/b cKO mice argue for the existence of a mitochondrial dysfunction at the level of the electron transport chain. Our results also suggest that Lmx1a/b cKO mitochondria operate with lower reserve capacity and thus might not cope effectively with periods of high bioenergetic demand. When overwhelmed, the mitochondrial respiratory chain will leak increased amounts of ROS (40). Even though the antioxidant system of neurons operates to minimize the detrimental effects of ROS, it may not be sufficient over time, and oxidative damage thus occurs. Oxidative stress can feed back to the mitochondria via a series of signaling pathways and cause further exacerbation and mitochondrial dysfunction by damaging mtDNA (Fig. 7). Our results demonstrating extensive mtDNA damage in Lmx1a/b cKO mice are in line with this model. High levels of mtDNA deletion have been reported in the SNpc of PD patients (41) and have been suggested to participate in disease pathogenesis (25).
Fig. 7.
A putative model of Lmx1a/b function in adult mDA neurons. Lmx1a/b control the expression of key genes involved in mitochondrial functions, and their ablation results in impaired respiratory chain activity, increased oxidative stress, and mitochondrial DNA damage. The predicted larger energetic requirements of mDA neurons make them more vulnerable to mitochondrial dysfunctions. Lmx1a/b deficiency caused axonal pathology characterized by α-synuclein+ inclusions, an autophagy defect followed by progressive loss of dopaminergic neurons.
A recent study reported that removal of Lmx1b in postnatal mDa neurons produced enlarged axon terminals associated with an alteration in autophagic flux (13). We also observed modification of the LC3-II protein level in 2-mo-old Lmx1a/b cKO mice, supporting the idea that the autophagic flux is altered in these mice. However, our mRNA-seq data in E15.5 tissue and RT-qPCR in P1 tissue did not reveal any significant change in the expression of autophagy–lysosomal pathway genes. Based on our results, it is likely that an alteration in mitochondrial metabolism occurs first and then leads to autophagy dysfunction (Fig. 7). Consistent with this notion, mitochondrial metabolism has been found to play a role in the modulation of autophagy (42). Because Lmx1a/b cKO mice recreate almost all the early cellular impairments associated with PD, this model could help us understand the respective contributions of mitochondrial dysfunction, α-synuclein aggregation/clearance, and autophagy in disease progression (Fig. 7).
A plethora of data argues for a central role of mitochondrial dysfunction in the pathogenesis of PD (43). The high vulnerability of DA neurons of the SNpc to degeneration is well known in PD but is not completely understood. New evidence favors the hypothesis that the selective vulnerability of SNpc DA neurons and other cell groups in PD can be explained in large part by these neurons’ high rate of basal mitochondrial oxidative metabolism and their particularly elaborate axonal arborization (34). The predicted larger energetic requirements in these neurons makes them more vulnerable to any further cellular stress induced by aging, environmental toxicants, or genetic risk factors. Lmx1a/b most likely contribute in coordinating the complex physiological demands of these neurons, both during development and in adulthood. Lmx1a/b mutant mice offer an unprecedented opportunity to investigate the cascade of events leading to the specific degeneration of mDA neurons and the links with mitochondrial dysfunctions.
In summary, our results provide insights into a novel Lmx1a/b-dependent pathway maintaining mitochondrial functions in midbrain dopaminergic neurons. Deletion of Lmx1a/b links metabolic impairment, α-synuclein inclusions, and progressive neuronal loss. Modulation of this pathway could rescue deficits and may have far-reaching implications for Parkinson’s disease.
Materials and Methods
More experimental details can be found in SI Materials and Methods.
Animals.
Lmx1a flox/flox; Lmx1b flox/flox mice have been described previously (7, 44) and were crossed with Dat-Cre heterozygous transgenic mice (15) to generate double-conditional Lmx1a/b mutants. We used this knock-in mouse line in which Cre recombinase expression is driven under the transcriptional control of the DAT promoter (15). Animals were housed and handled in accordance with the Canadian Guide for the Care and Use of Laboratory Animals. The Institutional Animal Care Committee of Laval University approved all surgical and animal care procedures.
Tissue Preparation and Immunohistochemistry.
Animals were anesthetized and perfused, and brains were removed. Postfixed and cryoprotected brains were snap frozen and cryosectioned in coronal sections (60 μm). Immunohistochemistry was performed on free-floating brain sections as described previously (45). All images were captured with a Zeiss LSM5 PASCAL confocal microscope or a Zeiss LSM 700 confocal microscope and were processed using Adobe Photoshop CS4.
Stereological Neuron Counting.
The number of TH+, DAT+, and Nissl+ neurons within the ventral midbrain was quantified using the optical fractionator stereological method (Stereo Investigator; MBF Bioscience) (46–48). The following analytic parameters were applied: 100 × 100 µm counting frame size; 48-µm optical dissector height; 1 in 3 section interval. Coefficients of error (Gundersen m = 1) were less than 0.07. The postnatal stages analyzed were P1, P20, P60, and P200 for TH and P60 for DAT and Nissl. Values are derived from at least four mice.
In Situ Hybridization.
Brains of wild-type (C57BL6) animals or Lmx1a/b cKO and control mice were fixed and removed. All solutions were treated with diethylpyrocarbonate. Tissue samples were cryosectioned in 20-μm coronal sections and mounted on Superfrost Plus slides (Fisher Scientific). The procedures for in situ hybridization have been described previously (8). The mouse anti-sense RNA probes for Lmx1a and Lmx1b were generated from cDNA templates using RT-PCR and were used as described previously (49). The cDNA template was generated from E18.5 whole-brain RNA.
Quantification of Dopamine and Metabolite Levels.
The concentration of dopamine and its metabolites [3,4-dihydroxyphenylacetic acid (DOPAC), 3-methoxytyramine (3-MT), and homovanillic acid (HVA)] were measured from the striatum of 20-d-old mice and mice more than 6 mo old. Sample preparation and quantification by HPLC with electrochemical detection were performed according to previously published protocols (50).
Metabolism Assays.
Primary midbrain cultures from Lmx1a/b cKO and control mice were prepared as described previously (51), with minor modifications. The rate of oxygen consumption deriving from mitochondrial oxidative phosphorylation was assessed using an extracellular flux analyzer (Seahorse Biosciences). After the assays, cells were immediately fixed for immunofluorescence. OCRs were normalized to a cell number identified with DAPI staining. Measurements made from wells containing only astrocytes were used to subtract the contribution of glial cells from the overall oxygen consumption of mixed neuron/astrocyte cultures. In a second step, the resulting values were normalized to the total number of neurons identified using MAP-2 immunostaining.
Western Blot.
Ventral midbrain or striatal lysates prepared in radioimmunoprecipitation assay (RIPA) buffer were separated by SDS/PAGE and transferred onto nitrocellulose membranes (Bio-Rad). TH, DDC, Lmx-1a, Lmx1b, α-synuclein, phospho-α-synuclein, and LC3 levels were assessed by Western blot analysis.
Rapid TH Immunostaining and LCM.
A combination of rapid TH immunochemistry and LCM was performed using P1 mice (21). A model AS-LMD LCM system (Leica Microsystems) was used to outline and capture the region of the sections with TH-immunoreactive neurons. RNA from microdissected tissue was isolated using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) according to the manufacturer’s protocol.
RNA Sequencing.
RNA was extracted using the PicoPure RNA Isolation Kit (Arcturus Engineering). The RNA-seq library was prepared using the Ovation RNA-Seq system (NuGEN). RNA-seq libraries were sequenced on an Illumina GAIIx, and analysis was performed using CLC Genomics Workbench version 1 (CLC bio). Expressed genes with an RPKM (reads per kilobase of transcript per million reads mapped) of 1 and above were described. Genes were described as differentially expressed when the expression difference of Lmx1a/b mutant tissue over control tissue was twofold or more. GO analysis was performed using the GOToolBox (genome.crg.es/GOToolBox), using the MGI identities of the list of genes. Gene lists and GO term results are provided in SI Materials and Methods.
RT-qPCR.
Real-time RT-qPCR analysis was performed using a LightCycler 480 (Roche Diagnostics) and analyzed with the LCS 480 software (version 1.5.0.39; Roche Applied Science). RNA samples obtained by LCM were reverse-transcribed into cDNA using the SuperScript III reverse transcriptase kit (Invitrogen). Relative quantifications were calculated using the ΔΔCT formula and normalizing the target gene amount over at least two or three reference genes (GAPDH, TBP, and RPL13). Data are presented as the mean ± SEM of the fold change relative to the expression at P1 and normalized against reference genes.
Stereotaxic Injection of Recombinant AAV.
AAV vectors expressing functional Cre-recombinase and nonfunctional Cre-recombinase (used as a control), both fused to GFP, were produced as described previously (52). See SI Materials and Methods for AAV vector generation. For stereotaxic injections, 2-mo-old Lmx1aF/F; Lmx1bF/F mice (n = 7) were used. To evaluate the effect of the age of ablation, 20-d-old and 20-mo-old Lmx1aF/F; Lmx1bF/F mice (n = 5) were used also. Mice were anesthetized with isoflurane, and skulls were immobilized in a stereotaxic apparatus. A 0.5-μL volume of virus [AAV Cre-GFP: 4.9 e10 genome copy (GC)/mL and AAV ΔCre-GFP: 8.6 e10 GC/mL] was injected unilaterally into the SNpc/VTA (anteroposterior: − 3.5 mm from bregma; mediolateral: 1.0 mm; dorsoventral: 4.0 mm). Animals were perfused 2, 4, or 6 mo after injection.
For the in vivo rescue experiment, 2-mo-old Lmx1aF/F; Lmx1bF/F mice were injected with a biscistronic vector coexpressing the functional Cre and Nrf1 (AAV Cre-GFP-Ires-Nrf1: 1.3 e12 GC/mL). Behavioral measurements were done at 2 and 6 mo after the injection. After behavioral measurements were performed, animals were perfused, and stereological analyses were done.
In addition to in situ hybridization (described above), β-gal staining and qualitative RT-PCR were used to confirm the efficiency of the viral vector (SI Materials and Methods and Fig. S6).
Locomotor Activity.
The locomotor activity of AAV-injected mice was tested 4 and 6 mo postinjection in locomotor detection chambers (AccuScan Instruments, Inc.) equipped with 16 light beams located in both the horizontal and vertical planes. The activity was monitored using the activity-monitoring VersaMax version 3.0 software (AccuScan Instruments, Inc.). Mice were allowed to habituate to the chamber for 15 min and were tested over a 60-min period in the open field. Activity was measured by recording horizontal and vertical activity and was quantified as the number of beam breaks per minute. Total distance traveled by an animal during 10 or 60 min was also measured as a locomotor activity parameter.
Detection of Mitochondrial DNA Damage.
The histochemical assay for the detection of abasic sites in the substantia nigra sections from 2-mo-old mice was performed as described previously (25). In brief, midbrain sections containing the SNpc and VTA were incubated with the ARP for 1 h, rinsed, and then immunostained for TH and the mitochondrial marker Tom20.
Culture of Primary DA Neurons, Transient Transfection, and ROS Assay.
Primary DA neuronal cultures were prepared from the mesencephalon of P1 mice. See SI Materials and Methods for the culture of primary DA neurons and details of transient transfection. In brief, after 1 d in vitro (DIV), cultures were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. After 3 DIV, transfected cells were either used for RNA extraction using an RNeasy Mini kit (Qiagen) and processed further for RT-qPCR (see above) or fixed and immunostained for TH. For the ROS assay, cultures were transfected with CMV-Nrf1 (Dharmacon) or (CMV-empty). ROS quantification was performed by epifluorescence microscopy using CellROX (Invitrogen) according to the manufacturer’s instructions on 3-d-old cultures (SI Materials and Methods). All images were captured with a Nikon Eclipse TE2000-U microscope and processed using MetaMorph, ImageJ, and Adobe Photoshop CS4.
Statistical Analysis.
All statistical analyses were performed with GraphPad Prism software 4.0 (GraphPad Software, Inc.). Data are expressed as mean ± SEM. The statistical significance of differences between two groups was calculated using Student's t test (with Welch's correction if the variance between the groups was different). Multiple groups of data were compared using ANOVA followed by the multiple comparisons test. Differences were considered significant at *P < 0.05; **P < 0.001; ***P < 0.0001.
SI Materials and Methods
Tissue Preparation and Immunohistochemistry.
Animals were deeply anesthetized with ketamine-xylazine (10 mg/mL and 1 mg/mL, respectively) and were transcardially perfused with PBS (pH 7.4) followed by 4% (wt/vol) formaldehyde in 100 mM phosphate buffer, pH 7.4. Brains were removed, immersion-fixed in the same fixative overnight at 4 °C, and subsequently cryoprotected in 30% (wt/vol) sucrose-PBS. Tissue samples were embedded in optimum cutting temperature (OCT) compound (Tissue-Tek) and cryosectioned into 60-μm-thick coronal sections. The slices were blocked for nonspecific antibody binding with PBS-BSA 1% and Triton 0.2% for 1 h and subsequently were incubated overnight at 4 °C with primary antibodies at optimized concentrations. The following primary antibodies were used: rabbit anti-TH (1:1,000; P40101-0; Pel-Freez Biologicals); sheep anti-TH (1:1,000; AB1542; Millipore); rabbit α-synuclein (1:100; 4179P; Cell Signaling Technology); anti–phospho-Ser129 (1:2,000; 015-25191; Wako Chemicals); rat anti-DAT (1:1,000; MAB369; Millipore); rabbit anti-Tom20 (1:500; sc-11415; Santa Cruz,); rabbit anti–LMX1a (1:1,000; AB10533; Millipore); and guinea pig anti-Lmx1b (1:10,000; a gift from Carmen Birchmeier, Max Delbrück Center for Molecular Medicine, Germany). Appropriate secondary antibodies conjugated to Cy3 (Jackson ImmunoResearch), FITC (Jackson ImmunoResearch), Alexa-Fluor 488 (711-545-152; Jackson ImmunoResearch), Alexa-Fluor 568 (A-11011; Life Technologies), Alexa-Fluor 647 (711-605-152; Jackson ImmunoResearch) or Northern Light 637 (NL011; R&D Systems) were used (1:200).
β-Gal Staining.
Sections were washed three times (10 min each washing) in 1× PBS + 0.02% Igepal (Nonidet P-40; Sigma-Aldrich) and then were incubated for 1 h at 37 °C in staining solution (1 mg/mL X-Gal (Invitrogen), 5 mM potassium ferricyanide (Sigma-Aldrich), 5 mM potassium ferrocyanide (Sigma-Aldrich), 5 mM EGTA (Bio-Basic), 0.01% sodium deoxycholate (Sigma-Aldrich), 2 mM MgCl2 (Sigma-Aldrich) and 1× PBS + 0.02% Nonidet P-40. The reaction was stopped with 1× PBS + 0.02% Nonidet P-40 when a blue precipitate appeared in the sections.
Nissl Staining.
Brain sections were mounted on slides and initially soaked in 70% (vol/vol) alcohol for 10 min followed by 5 min in water. Slides were then submerged in 1.5% (wt/vol) Nissl staining solution. Finally, slides were soaked in water with increasing concentrations (70, 95, and 100%) of alcohol followed by soaking in toluene.
Quantification of Dopamine and Metabolite Levels.
Mice were decapitated, and their brains were quickly dissected and snap frozen in liquid nitrogen. HPLC quantification was normalized to protein concentrations. Protein measurements were determined with a detergent-compatible (DC) protein assay kit (Bio-Rad) using BSA as a standard according to the manufacturer’s protocol.
Metabolism Assays.
Primary midbrain cultures from Lmx1a/b cKO and control mice were prepared as described previously (51) with minor modifications. After dissection, midbrain neurons were dissociated using papain (Worthington) and seeded on a monolayer of cortical astrocytes grown on poly-l-lysine (Sigma)–coated glass coverslips. The total seeding density was 250,000 cells/mL. All cell cultures were grown at 37° in 5% CO2 and maintained in Neurobasal (Life Technologies) and minimum essential medium containing 1% penicillin/streptomycin, 1% GlutaMAX, 2% (vol/vol) B-27 supplement, and 5% (vol/vol) FBS (Invitrogen). The OCR deriving from mitochondrial oxidative phosphorylation was assessed using an extracellular flux analyzer (Seahorse Biosciences). Cells were plated on XF24 tissue-culture plates and maintained in culture for 10 d. Before the experiments, cells were incubated for 1 h at 37 °C in a CO2-free incubator in bicarbonate-free DMEM (Sigma) supplemented with 200 mM GlutaMAX-1 (Invitrogen), 1 mM sodium pyruvate (Sigma), 25 mM d-glucose (Sigma), 63.3 mM NaCl (Sigma), and phenol red (Sigma). The pH was adjusted to 7.4 with NaOH. Oxygen consumption was measured sequentially under basal conditions in the presence of the mitochondrial uncoupler CCCP (0.5 μM) and the mitochondrial inhibitors rotenone (1 μM) and antimycin A (1 μM) to assess the maximal oxidative capacity and nonmitochondrial oxygen consumption, respectively.
Western Blot.
Control and Lmx1a/b cKO mice at different ages were anesthetized and decapitated. The brains were quickly dissected out and snap frozen in liquid nitrogen. Striatal sections of 60-μm thickness were cut using a cryostat. Striatal section lysates or the dissected ventral midbrain were extracted in RIPA buffer containing protease inhibitor and phosphatase inhibitor mixtures (Roche) (50 mM Tris⋅HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 50 mM NaF, pH 7.4) and were boiled for 5 min. The protein concentration was measured using a DC-protein assay (Bio-Rad). Protein extracts (10 μg) were separated on 4–20% (vol/vol) SDS/PAGE Tris-glycine gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). Blots were immunostained overnight at 4 °C with primary antibodies. The primary antibodies actin (1:20,000; MAB1501; Millipore), TH (1:1,000; P40101-0; Pel-Freez), DDC (1:750; AB1569; Millipore), Lmx1a (1:1,000; ab31006; Abcam), Lmx1b (1:500; AP8935c; Abgent), α-synuclein (1:1,000; 4179; Cell Signaling Technology), phospho-α-synuclein (1:1,000; ab51253; Abcam), and LC3 (1:500; AP1801a; Abgent) were diluted in blocking solution containing 1× PBS and 0.01% Tween-20 (Sigma). Blots were washed three times (10 min each washing) with 1× PBS and 0.01% Tween-20, and immune complexes were detected with species-appropriate secondary antibody conjugated to HRP: goat anti-rabbit HRP (1:3,000; 7074; CST) and goat anti-mouse HRP (1:5,000; G-21040; Life Technologies). The chemiluminescence signal (Western Lightning Plus-ECL; PerkinElmer) was exposed to Pierce CL-Xposure film (Thermo Scientific). Films were scanned and analyzed using the ImageJ64 program.
Generation of AAV Vectors.
Recombinant AAV Cre-GFP, AAV ΔCre-GFP, and AAV Cre-GFP-Ires-Nrf1 vectors used in this study were prepared by the Canadian neurophotonics platform viral vector core. Briefly, genes were amplified by PCR using the Q5 enzyme (New England Biolabs) and assembled into plasmids for the generation of AAV vectors using Gibson Assembly (New England Biolabs). Correct clones for each vector were amplified in Stbl2 Escherichia coli (Invitrogen), and the DNA was purified in endotoxin-free conditions (Qiagen). AAV2 pseudotyped with the AAV1 capsid were produced and purified. Briefly, 293T17 cells were cotransfected with a plasmid containing helper genes, a plasmid containing Rep/Cap, and the plasmids containing Cre-GFP, ΔCre-GFP, or Cre-GFP-Ires-Nrf1. Cells were harvested 48 h after transfection, and viral particles were released through four freeze-thaw cycles. Free DNA was digested with Benzonase, and debris was centrifugated out. Viral particles were purified by a discontinuous gradient of iodixanol [15, 25, 40, and 60% (vol/vol)] and ultracentrifugation (504,300 × g for 90 min at 16 °C). The viral preparation was concentrated through a Centricon filter (Millipore) and adjusted to 5% (wt/vol) sorbitol for storage and experimentation. Titration was performed qPCR using primers specific for the CAG promoter and several dilutions of the viral preparation. The plasmids encoding the corresponding gene were used as controls.
Rapid TH Immunostaining and LCM.
The entire VTA and SNpc were dissected from midbrain sections stained using a previously described protocol (1). Briefly, tissue sections mounted onto LCM membrane slides (Leica Biosystems) were fixed in cold ethanol, incubated with rabbit anti-TH, and exposed to biotinylated anti-rabbit antibody. The slides were subsequently incubated in avidin/biotinylated enzyme complex-HRP (Vectastain; Vector Laboratories), and the staining was detected with the diaminobenzidine substrate. Sections were dehydrated in absolute ethanol and air-dried. The entire VTA and SNpc were dissected from midbrain sections using a Leica laser microdissection system (Model AS-LMD). Samples were collected in lysis buffer, and mRNA was extracted using an RNA isolation kit (Arcturus PicoPure RNA Isolation Kit).
RT-qPCR.
Real-time PCR reactions were set up in a volume of 20 μL using the FastStart Essential DNA Green Master Kit (Roche Applied Science), and amplification was performed using the SYBR Green 96-well program (50 °C for 2 min; 95 °C for 15 min; 40 amplification cycles at 94 °C for 15 s and 60 °C for 1 min). Primers were designed in the 3′ region of each gene using Primer3 software (53). The primer sequences are available upon request. Each reaction was carried out in triplicate. Relative gene expression was estimated using the Advanced Relative Quantification mode in the LCS 480 software. Melting-curve analysis was also performed to confirm homogeneous product formation, without primer-dimers or nonspecific product amplifications.
Culture of Primary DA Neurons and Transient Transfection.
The ventral mesencephalon of P1 mouse embryos were dissected in L-15 medium (Life Technologies) and dissociated by papain treatment [12 U/mL papain (Worthington Biochemical); 250 U/mL DNase I type IV (Sigma); 3.5 mM l-cysteine (Sigma-Aldrich); 0.215% NaHCO3 (Sigma-Aldrich); 5 mM EDTA (Life Technologies); 0.2% Phenol Red (Sigma-Aldrich); 1 mM sodium pyruvate (Life Technologies); 1.8 mg/mL d-glucose (Sigma-Aldrich); 50 U/mL penicillin and 50 μg/mL streptomycin (Life Technologies) in HBSS without Ca2+ and Mg2+ (Life Technologies)], before mechanical trituration in trituration solution [0.2% BSA (Sigma-Aldrich); 50 U/mL penicillin, 50 μg/mL streptomycin; 1 mM sodium pyruvate; 1.8 mg/mL glucose; 250 U/mL DNase I type IV in Neurobasal medium (Life Technologies)]. Cells were purified on BSA columns [1.8% (wt/vol) BSA; 50 U/mL penicillin; 50 μg/mL streptomycin; 1 mM sodium pyruvate; 1.8 mg/mL glucose; 250 U/mL DNase I type IV; 3 mM NaOH (Fisher Scientific) in Neurobasal medium] at 800 × g for 5 min. Cells were seeded on eight-well culture slides (Fisher Scientific) or 12-mm coverslips (Fisher Scientific) coated with 0.003% poly-l-ornithine (Sigma) and 10 μg/mL laminin (Life Technologies) and were maintained at 37 °C in a humidified atmosphere of 5% CO2 in complete growth medium [10% (vol/vol) FBS (Life Technologies); 1:50 B27 supplements (Life Technologies); 1:100 GlutaMAX (Life Technologies); 1.2 mg/mL d-glucose in Neurobasal medium] at low density (3,000,000 cells/mL). Cells were transfected at DIV 2 with the appropriate plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Caggs-Lmx1a-Ires-EGFP was a gift from S.-L.A. CMV-Nrf1 came from GE Dharmacon, and CMV-Lmx1b-RFP came from ABM. The control plasmids CMV-empty, CMV-Ires-tdTomato, CMV-mCherry, and CMV-EGFP came from Plateforme d’Outils Moléculaires, Institut Universitaire en Santé Mentale de Québec.
ROS Assay.
To quantify the ROS-sensitive probe signal in neurons, a single in-focus plane (phase-contrast and fluorescence) was acquired. Ten neurons per condition (mouse genotype × transfected vector) were randomly selected, and CTCF was calculated using the formula: CTCF = integrated density − (area of selected cell × mean fluorescence of background readings), as described in ref. 54. Five to seven mice of each genotype were analyzed in at least three independent experiments (Fig. 6 A and B).
In addition, the CTCF was also calculated in transfected and TH+ neurons (Fig. S9A). For this experiment, cultures were cotransfected with CMV-Nrf1 (Dharmacon) or (CMV-empty) and CMV-mCherry. At 3 DIV, cells were fixed and immunostained for TH. ROS quantification was then performed as described in Materials and Methods.
Qualitative RT-PCR.
Lmx1a/bF/F mice that received a unilateral injection of AAV ΔCre-GFP, AAV Cre-GFP, or AAV Cre-GFP-Ires-Nrf1 in SNpc were killed 2 mo after injection. Brains were removed immediately, snap-frozen, placed on a brain slicer matrix (Zivic Instruments), and sliced into 1-mm-thick coronal sections at the injection level. Brains were also hemisected to divide the sections into left and right hemispheres. Total RNA was extracted using the RNeasy mini kit (Qiagen) followed by DNase-I treatment, according to the manufacturer’s instructions. Reverse transcription was carried out using SuperScript III (Invitrogen) according to the manufacturer’s instructions. PCR amplification was performed using the single-stranded cDNA as a template with primers designed to target Lmx1a and Lmx1b transcripts. PCR products were separated by 2% (wt/vol) agarose gel electrophoresis.
Acknowledgments
We thank Drs. Armen Saghatelyan, Simon Stott, and Emmanouil Matzakopian for useful comments and suggestions; Dr. Carmen Birchmeier for sharing Lmx1b antibody; Richard D. Palmiter for Cre-GFP and ΔCre-GFP plasmids; and the Plateforme d’Outils Moléculaire (https://neurophotonics.ca/fr/pom) for the production of viral vectors. This work was funded by Canadian Institutes of Health Research Grants MOP 311120 (to M.L.) and MOP106556 (to L.-E.T.) and by Natural Sciences and Engineering Research Council of Canada Grant 418391-2012 (to M.L.). H.D.-B. received a fellowship from the Fonds du Québec en Recherche, Santé (FRQS) and Fonds Québécois de Recherche sur le Parkinson. M.L. received salary support from the FRQS. N.G. and C.P. receive salary support from Parkinson Society Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520387113/-/DCSupplemental.
References
- 1.Shulman JM, De Jager PL, Feany MB. Parkinson’s disease: Genetics and pathogenesis. Annu Rev Pathol. 2011;6(1):193–222. doi: 10.1146/annurev-pathol-011110-130242. [DOI] [PubMed] [Google Scholar]
- 2.Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat Rev Neurosci. 2006;7(3):207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- 3.Blaess S, Ang SL. Genetic control of midbrain dopaminergic neuron development. Wiley Interdiscip Rev Dev Biol. 2015;4(2):113–134. doi: 10.1002/wdev.169. [DOI] [PubMed] [Google Scholar]
- 4.Hegarty SV, Sullivan AM, O’Keeffe GW. Midbrain dopaminergic neurons: A review of the molecular circuitry that regulates their development. Dev Biol. 2013;379(2):123–138. doi: 10.1016/j.ydbio.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Doucet-Beaupré H, Lévesque M. 2013. The role of developmental transcription factors in adult midbrain dopaminergic neurons. OA Neurosciences 1(1):3.
- 6.Smidt MP, et al. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000;3(4):337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 7.Deng Q, et al. Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development. 2011;138(16):3399–3408. doi: 10.1242/dev.065482. [DOI] [PubMed] [Google Scholar]
- 8.Yan CH, Levesque M, Claxton S, Johnson RL, Ang SL. Lmx1a and lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J Neurosci. 2011;31(35):12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16(2):75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 10.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476(7359):224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 11.Bergman O, et al. Do polymorphisms in transcription factors LMX1A and LMX1B influence the risk for Parkinson’s disease? J Neural Transm (Vienna) 2009;116(3):333–338. doi: 10.1007/s00702-009-0187-z. [DOI] [PubMed] [Google Scholar]
- 12.Dai JX, Hu ZL, Shi M, Guo C, Ding YQ. Postnatal ontogeny of the transcription factor Lmx1b in the mouse central nervous system. J Comp Neurol. 2008;509(4):341–355. doi: 10.1002/cne.21759. [DOI] [PubMed] [Google Scholar]
- 13.Laguna A, et al. Dopaminergic control of autophagic-lysosomal function implicates Lmx1b in Parkinson’s disease. Nat Neurosci. 2015;18(6):826–835. doi: 10.1038/nn.4004. [DOI] [PubMed] [Google Scholar]
- 14.Zou HL, et al. Expression of the LIM-homeodomain gene Lmx1a in the postnatal mouse central nervous system. Brain Res Bull. 2009;78(6):306–312. doi: 10.1016/j.brainresbull.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang X, Masson J, Gingrich JA, Rayport S, Hen R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. J Neurosci Methods. 2005;143(1):27–32. doi: 10.1016/j.jneumeth.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 16.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1(1):4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huot P, et al. L-Dopa treatment abolishes the numerical increase in striatal dopaminergic neurons in parkinsonian monkeys. J Chem Neuroanat. 2008;35(1):77–84. doi: 10.1016/j.jchemneu.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Porritt MJ, et al. New dopaminergic neurons in Parkinson’s disease striatum. Lancet. 2000;356(9223):44–45. doi: 10.1016/S0140-6736(00)02437-5. [DOI] [PubMed] [Google Scholar]
- 19.Hoekstra EJ, et al. Lmx1a is an activator of Rgs4 and Grb10 and is responsible for the correct specification of rostral and medial mdDA neurons. Eur J Neurosci. 2013;37(1):23–32. doi: 10.1111/ejn.12022. [DOI] [PubMed] [Google Scholar]
- 20.Hoekstra EJ, Mesman S, de Munnik WA, Smidt MP. LMX1B is part of a transcriptional complex with PSPC1 and PSF. PLoS One. 2013;8(1):e53122. doi: 10.1371/journal.pone.0053122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabrat A, Doucet-Beaupré H, Lévesque M. 2015. RNA isolation from cell specific subpopulations using laser-capture microdissection combined with rapid immunolabeling. Journal of Visualized Experiments 98:e52510. [DOI] [PMC free article] [PubMed]
- 22.Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF, and intron Sp1 recognition sequences. J Biol Chem. 1989;264(24):14361–14368. [PubMed] [Google Scholar]
- 23.Pemberton S, et al. Hsc70 protein interaction with soluble and fibrillar alpha-synuclein. J Biol Chem. 2011;286(40):34690–34699. doi: 10.1074/jbc.M111.261321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pemberton S, Melki R. The interaction of Hsc70 protein with fibrillar α-Synuclein and its therapeutic potential in Parkinson’s disease. Commun Integr Biol. 2012;5(1):94–95. doi: 10.4161/cib.18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders LH, et al. Mitochondrial DNA damage: Molecular marker of vulnerable nigral neurons in Parkinson’s disease. Neurobiol Dis. 2014;70(0):214–223. doi: 10.1016/j.nbd.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goedert M, Spillantini MG, Del Tredici K, Braak H. 100 years of Lewy pathology. Nat Rev Neurol. 2013;9(1):13–24. doi: 10.1038/nrneurol.2012.242. [DOI] [PubMed] [Google Scholar]
- 27.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24(1):117–129. doi: 10.1016/s1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 28.Yim YS, et al. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc Natl Acad Sci USA. 2013;110(10):4057–4062. doi: 10.1073/pnas.1209881110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proenca CC, Gao KP, Shmelkov SV, Rafii S, Lee FS. Slitrks as emerging candidate genes involved in neuropsychiatric disorders. Trends Neurosci. 2011;34(3):143–153. doi: 10.1016/j.tins.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke RE, O’Malley K. Axon degeneration in Parkinson’s disease. Exp Neurol. 2013;246(0):72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu Y, et al. Alterations in axonal transport motor proteins in sporadic and experimental Parkinson’s disease. Brain. 2012;135(Pt 7):2058–2073. doi: 10.1093/brain/aws133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galter D, et al. MitoPark mice mirror the slow progression of key symptoms and L-DOPA response in Parkinson’s disease. Genes Brain Behav. 2010;9(2):173–181. doi: 10.1111/j.1601-183X.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, et al. Cognitive dysfunction precedes the onset of motor symptoms in the MitoPark mouse model of Parkinson’s disease. PLoS One. 2013;8(8):e71341. doi: 10.1371/journal.pone.0071341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacelli C, et al. Elevated mitochondrial bioenergetics and axonal arborization size are key contributors to the vulnerability of dopamine neurons. Curr Biol. 2015;25(18):2349–2360. doi: 10.1016/j.cub.2015.07.050. [DOI] [PubMed] [Google Scholar]
- 35.Greenamyre JT, Sherer TB, Betarbet R, Panov AV. Complex I and Parkinson’s disease. IUBMB Life. 2001;52(3-5):135–141. doi: 10.1080/15216540152845939. [DOI] [PubMed] [Google Scholar]
- 36.Rak M, Rustin P. Supernumerary subunits NDUFA3, NDUFA5 and NDUFA12 are required for the formation of the extramembrane arm of human mitochondrial complex I. FEBS Lett. 2014;588(9):1832–1838. doi: 10.1016/j.febslet.2014.03.046. [DOI] [PubMed] [Google Scholar]
- 37.Dhar SS, Ongwijitwat S, Wong-Riley MTT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283(6):3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong-Riley MT. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv Exp Med Biol. 2012;748:283–304. doi: 10.1007/978-1-4614-3573-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satoh J, Kawana N, Yamamoto Y. Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Bio. 2013;7:139–152. doi: 10.4137/GRSB.S13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38(5):518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 42.Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell. 2014;159(6):1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Exner N, Lutz AK, Haass C, Winklhofer KF. Mitochondrial dysfunction in Parkinson’s disease: Molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31(14):3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Z-Q, et al. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J Neurosci. 2006;26(49):12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferri AL, et al. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134(15):2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 46.Ma J, Shaw VE, Mitrofanis J. Does melatonin help save dopaminergic cells in MPTP-treated mice? Parkinsonism Relat Disord. 2009;15(4):307–314. doi: 10.1016/j.parkreldis.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Peoples C, et al. Photobiomodulation enhances nigral dopaminergic cell survival in a chronic MPTP mouse model of Parkinson’s disease. Parkinsonism Relat Disord. 2012;18(5):469–476. doi: 10.1016/j.parkreldis.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Wallace BA, et al. Survival of midbrain dopaminergic cells after lesion or deep brain stimulation of the subthalamic nucleus in MPTP-treated monkeys. Brain. 2007;130(Pt 8):2129–2145. doi: 10.1093/brain/awm137. [DOI] [PubMed] [Google Scholar]
- 49.Krawchuk D, Kania A. Identification of genes controlled by LMX1B in the developing mouse limb bud. Dev Dyn. 2008;237(4):1183–1192. doi: 10.1002/dvdy.21514. [DOI] [PubMed] [Google Scholar]
- 50.Bousquet M, et al. Transgenic conversion of omega-6 into omega-3 fatty acids in a mouse model of Parkinson’s disease. J Lipid Res. 2011;52(2):263–271. doi: 10.1194/jlr.M011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fasano C, Thibault D, Trudeau LE. 2008. Culture of postnatal mesencephalic dopamine neurons on an astrocyte monolayer. Curr Protoc Neurosci Chapter 3:Unit 3 21.
- 52.Zweifel LS, Argilli E, Bonci A, Palmiter RD. Role of NMDA receptors in dopamine neurons for plasticity and addictive behaviors. Neuron. 2008;59(3):486–496. doi: 10.1016/j.neuron.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz S, editors. Bioinformatics Methods and Protocols, Methods in Molecular Biology. Vol 132. Humana Press; New York: 1999. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 54.McCloy RA, et al. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13(9):1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]