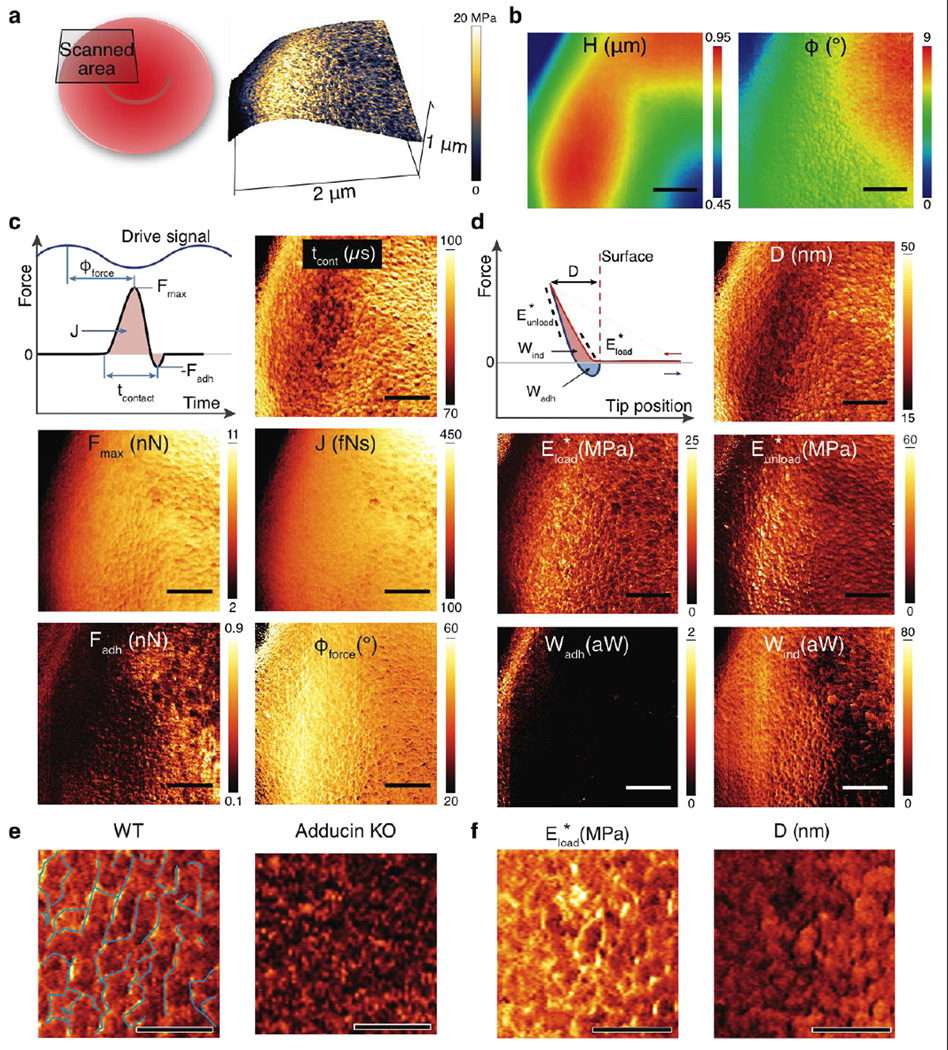
Figure 2. Nanomechanical images showing spectrin networks of a red blood cell (RBC).
(a) 3D Topography showing ~0.5 µm variation across the image and the concaved center of the RBC. The coloration of the 3D surface was from the reduced Young’s modulus image. The side–wall of RBC (the left edge) was imaged at a shallow angle, resulting in skewed measurements of nanomechanical properties. (b) Conventional topography (H) and phase(φ0) images (256 × 256 pixels) as in tapping–mode AFM. (c) Images of nanomechanical properties directly measured from the time–resolved tip–sample interaction force: tip–cell contact time (tcont), peak repulsive and adhesive forces (Fmax and Fadh), impulse (J, force integrated over time) and force phase (φforce, phase lag of the peak repulsive force to the driven base motion of the cantilever). (d) Images of nanomechanical properties derived from reconstructed force–distance curves using calculated indentation: maximal indentation (D), reduced Young’s moduli (stiffness) from the first 25 nm of loading and the upper half of the unloading force curve (E*load and E*unload, respectively), and energy dissipation due to indentation (Wind) and adhesion (Wadh). The nanomechanical images provide quantitative measures of local mechanical properties with a large number of force probing (25 taps per pixel). Scale bar is 500 nm and image size is 841 × 256 pixels. (e) Zoomed in map of elastic modulus of an RBC from a WT and Adducin KO mouse. Scale bar is 200 nm, color scale is identical to Figure 2d. The polygonal spectrin network is shown by superimposed, thin blue lines. (f) Zoomed in maps of elastic modulus and indentation depth show stark differences in the region of the spectrin network versus non–spectrin regions. Scale bar is 200 nm, color scale is identical to Figure 2d.