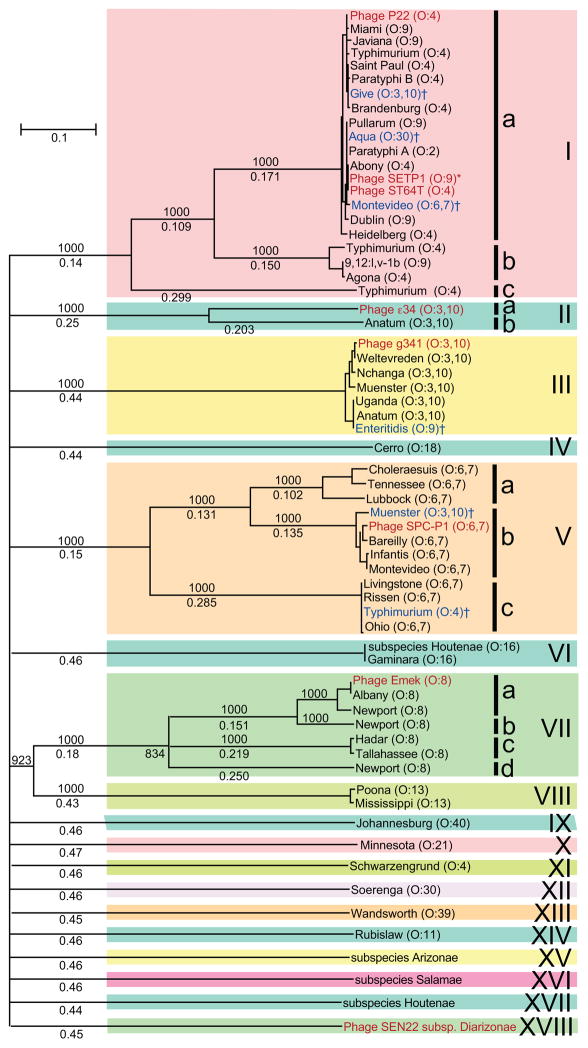
Figure 7. Neighbor-joining tree of Salmonella P22-like tailspikes.
Receptor binding domains encoded by P22-like phage and prophage tailspikes were aligned, and an unrooted tree was constructed with ClustalX (Larkin et al., 2007) Tailspike locus_tags are listed in Table S4H. Selected bootstrap values (out of 1000 trials) are shown above the branch lines, and fractional difference lengths are indicated below the lines; long branches with bootstrap values less than 800 were collapsed. Reported serotypes of the prophage host strains and O group (in parentheses) are indicated at the tip of each branch; blue text indicates the “anomalous” tailspikes discussed in the text, and daggers (†) denote O groups that are probably incorrectly determined (see text and Figure S15); red text denotes authentic phage tailspikes and the asterisk (*) denotes a tailspike from the incompletely sequenced S. enterica serovar Enteritidis P22-like phage SETP1 (De Lappe et al., 2009). Colored boxes highlight the eighteen different tailspike types with their names given at the right.