Figure 1. The LD protein Pgc1 is an ERAD substrate.
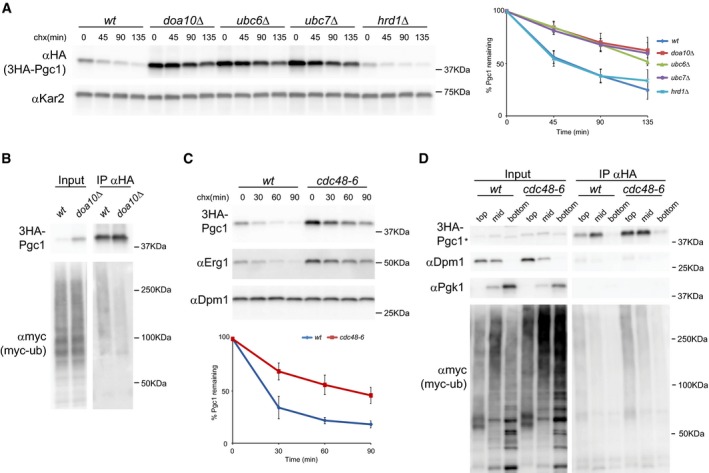
- The degradation of 3HA‐Pgc1 was analyzed in cells with the indicated genotype upon inhibition of protein synthesis with cycloheximide (chx). A plasmid‐borne 3HA‐Pgc1 expressed from the endogenous promoter was used. 3HA‐Pgc1 was detected with anti‐HA antibodies. Kar2 was used as a loading control and detected with anti‐Kar2 antibodies. The graph on the right shows the average of four independent experiments; error bars represent the standard deviation.
- The ubiquitination of 3HA‐Pgc1 was analyzed in cells with the indicated genotype and expressing myc‐tagged ubiquitin. 3HA‐Pgc1 was immunoprecipitated using anti‐HA and polyubiquitin‐conjugated 3HA‐Pgc1 was detected using anti‐myc antibody. Input fraction corresponds to 2% of the total amount used for IP.
- The degradation of 3HA‐Pgc1 was analyzed as in (A) in cells bearing the CDC48 temperature‐sensitive allele cdc48‐6. Cells were grown 2.5 h at 37°C prior to addition of cycloheximide. Inactivation of Cdc48 mutant protein was confirmed by stabilization of the ERAD substrate Erg1 in the same cells. The graph shows the average of four independent experiments; error bars represent the standard deviation.
- Membrane association of ubiquitinated 3HA‐Pgc1 was analyzed by density gradient centrifugation followed by immunoprecipitation. Cells of the indicated genotype and expressing myc‐tagged ubiquitin were grown as in (C). Lysates were subjected to centrifugation on an Optiprep density gradient. Collected fractions were analyzed by Western blotting before and after immunoprecipitation with anti‐HA antibodies. Dpm1 and Pgk1 were used as markers for membrane‐bound and soluble fractions, respectively. Input fraction corresponds to 10% of the total amount used for IP. “Top”, “mid”, and “bottom” indicate the fractions from the gradient. Asterisk marks a faster migrating unspecific band in the input bottom fraction.
Source data are available online for this figure.
