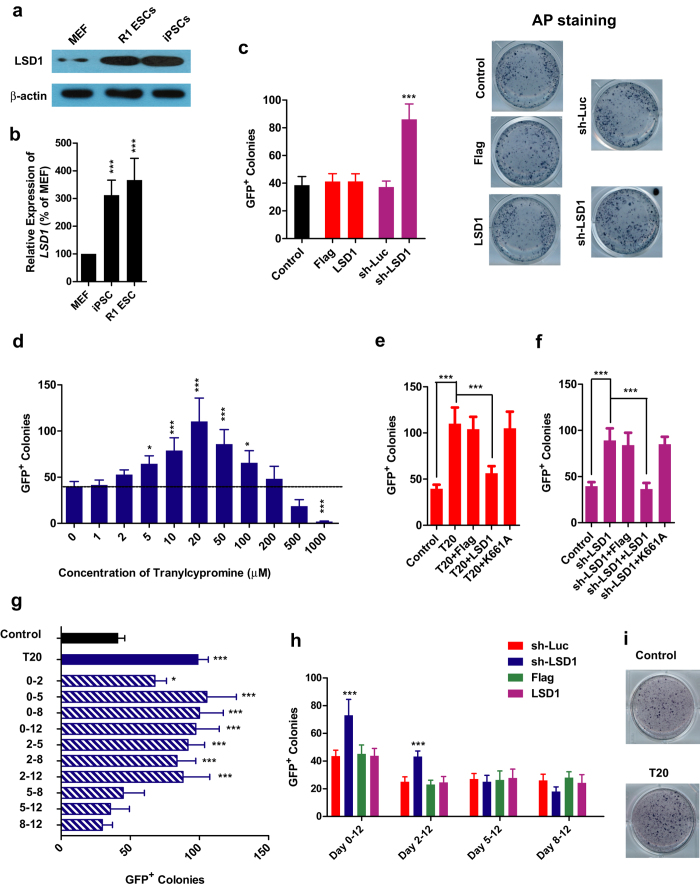
Figure 1. LSD1 inhibition promotes iPSCs generation.
(a,b) Lsd1 expression was determined with immunoblotting ((a), n = 5) and qPCR ((b), n = 5) in MEFs, iPSCs and R1 ESCs. (c) Reprogramming was performed with Lsd1 up- and down-regulation. GFP+ colonies numbers and AP staining results were summarized (n = 6). (d) Different concentrations of LSD1 inhibitor, tranylcypromine, were used for reprogramming. Efficiencies were evaluated by counting GFP+ colonies (n = 6). (e,f) 20 μM tranylcypromine (T20), retrovirus encoding Flag vector, LSD1, LSD1 deficient mutant, K661A, and shRNA against Lsd1 (sh-LSD1) were used for reprogramming as indicated. GFP+ colonies were counted (n = 6). (g) T20 was used during indicated periods during reprogramming, and GFP+ colonies were counted (n = 6). (h) Retrovirus encoding Flag vector, Lsd1, shRNA against Luciferese (sh-Luc) and sh-LSD1 were delivered at different time points during reprogramming before counting GFP+ colonies on Day 12 (n = 6). (i) T20 was used for HFFs reprogramming. AP staining was performed to determining the efficiencies (n = 5). One-way ANOVA with Dunnett’s test as a post-hoc was used to compare the experimental with control groups. The gels used for different antibodies were run under the same experimental conditions (a). Representative images were cropped from the original images with no modification on the relative intensities. The full length blots were provided in Supplementary Figure S8.