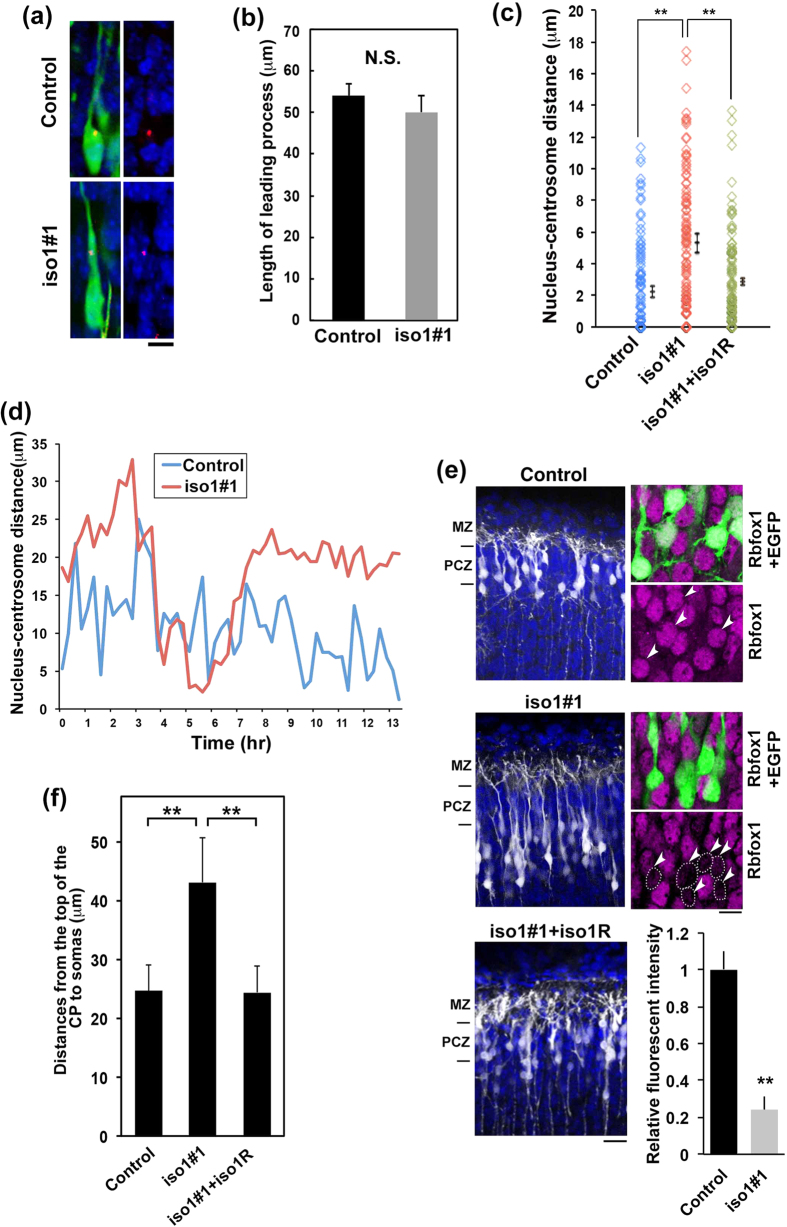
Figure 6. Coupling of the nucleus to centrosome is defective in Rbfox1-iso1-deficient neurons.
(a) Cell shape of control and Rbfox1-iso1-deficient neurons during radial migration. pCAG-EGFP was electroporated with pCAG-PACKmKO1 (a marker for centrosome) with pSuper-H1.shLuc or pSuper-mRbfox1-iso1#1 into cerebral cortices at E14.5. GFP, centrosome (red) and DNA (blue) were immunostained in coronal sections at E17.5. Representative images of migrating neurons in lower CP were shown. Scale bar, 10 μm. (b) Quantification of the length of leading process of control and Rbfox1-iso1-deficient neurons. Numbers of cells for each calculation was more than 50. Error bars indicate SD. (c) The N-C distance between centrosome and the top of nucleus was measured. pCAG-EGFP was transfected with pCAG-PACKmKO1 together with pSuper-H1.shLuc (Control), pSuper-mRbfox1-iso1#1 or pSuper-mRbfox1-iso1#1 plus pCAG-Myc-mRbfox1-iso1R into E14.5 mouse brains, and fixed at E17.5. Numbers of cells for each calculation was 100 cells. Error bars indicate SD; **p < 0.01 by Tukey-Kramer LSD (n = 3). (d) Time-course profiles of the N-C distance dynamics of control and the deficient neurons (iso1#1). (e) Effects of Rbfox1-iso1-knockdown for the terminal translocation. Cerebral cortices were electroporated with pCAG-EGFP with pSuper-H1.shLuc (Control), pSuper-mRbfox1-iso1#1 or pSuper-mRbfox1-iso1#1 plus pCAG-Myc-mRbfox1-iso1R at E15.5, and analyzed at P3. MZ, marginal zone; PCZ, primitive cortical zone. Rbfox1 expression was also analyzed (right panels). GFP-positive cells were indicated by arrowheads while Rbfox1-deficient cells were encircled by dotted line. Quantification of Rbfox1 expression was performed for the deficient cells as in Fig. 2e. (f) Statistical analyses of (e). Distance between the top of CP and the cell soma was measured. Error bars represent SD. **p < 0.01 by Tukey-Kramer LSD (n = 3).