Fig. 5.
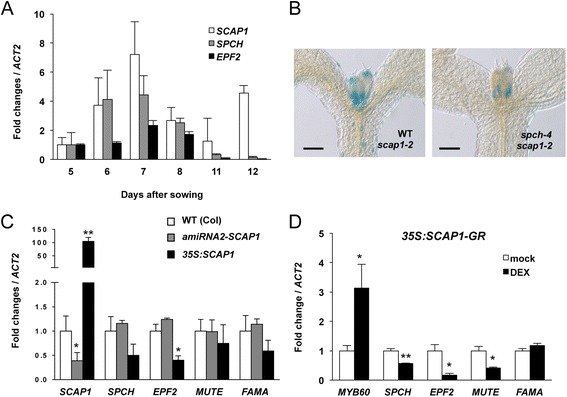
Role of SCAP1 on stomatal genes transcript accumulations. (a) Pattern of SCAP1, SPCH and EPF2 transcript accumulations determined by quantitative PCR in manually dissected first two leaf primordia of wild type (Col) seedlings at different days after sowing. Values represent the mean of three biological replicates (30 leaves / replica). (b) GUS staining of scap1-2 in wild type or spch-4 mutant background in 5 day old seedlings. Bar = 100 μm. (c) Pattern of SCAP1, SPCH, EPF2, MUTE and FAMA transcript accumulations determined by quantitative PCR in manually dissected first two leaf primordia of 7 days-old wild type (Col), pro35S:amiRNA2-SCAP1 (amiRNA2-SCAP1) and pro35S:SCAP1-YFP (35S:SCAP1) plants. Values represent the mean of three biological replicates (30 leaves / replica). (d) Pattern of AtMYB60, SPCH, EPF2, MUTE and FAMA transcript accumulations determined by quantitative PCR in 10 days-old pro35S:SCAP1-GR (35S:SCAP1-GR) plants treated by spraying with DEX (or mock) and the whole seedlings were sampled at eight hours after treatment. Values represent the mean of two biological replicates. In all quantitative PCR ACTIN (ACT2) was used for normalization. In c and d ** = P < 0.01 and * = P < 0.05 and two tails T Student test. Error bars = standard deviation
