Abstract
Arsenic (+3 oxidation state) methyltransferase (AS3MT) is the key enzyme in the metabolism of inorganic arsenic (iAs). Polymorphisms of AS3MT influence adverse health effects in adults, but little is known about their role in iAs metabolism in pregnant women and infants. The relationships between seven single nucleotide polymorphisms (SNPs) in AS3MT and urinary concentrations of iAs and its methylated metabolites were assessed in mother-infant pairs of the Biomarkers of Exposure to ARsenic (BEAR) cohort. Maternal alleles for five of the seven SNPs (rs7085104, rs3740400, rs3740393, rs3740390, and rs1046778) were associated with urinary concentrations of iAs metabolites, and alleles for one SNP (rs3740393) were associated with birth outcomes/measures. These associations were strongly dependent upon the male sex of the fetus but independent of fetal genotype for AS3MT. These data highlight a potential sex-dependence of the relationships among maternal genotype, iAs metabolism and infant health outcomes.
Keywords: AS3MT, arsenic, genotype, prenatal exposure, birth outcomes
1. Introduction
Inorganic arsenic (iAs) is a developmental toxicant that crosses the placenta reaching fetal organs [1–3]. Exposure to iAs during pregnancy has been associated with adverse health and infant outcomes including increased risk of spontaneous abortion, stillbirth, infant mortality, low birth weight, decreased head and chest circumferences, and increased risk of infection in infants [4]. In addition to being associated with adverse outcomes in early life, prenatal and early childhood exposures to iAs have been associated with higher rates of mortality in adulthood [5]. There is increasing information that the detrimental health impacts of prenatal iAs exposure are associated not only with levels of exposure to iAs but also its biotransformation.
In humans, ingested iAs is biotransformed via multiple consecutive methylation steps into the methylated metabolites, monomethyl-arsenic (MMAs) and dimethyl-arsenic (DMAs), a process that has been described as enhancing removal and thus decreasing the overall toxicity of arsenic [6]. Both inorganic and methylated arsenicals have been detected in trivalent (AsIII) and pentavalent (AsV) oxidation states in the urine of subjects chronically exposed to arsenic [6, 7]. Notably, trivalent MMAsIII and DMAsIII have been shown to be more cytotoxic, genotoxic and potent inhibitors of some enzymes than their pentavalent counterparts [8], thus making them critical intermediate metabolites of adverse effects associated with iAs exposure. The urinary levels of excreted iAs and its metabolites are used to assess individual arsenic exposure and methylation efficiency. In general, individuals who are thought to be efficient in iAs methylation have lower percentages of total urinary arsenic present as iAs (10–20%) and MMAs (10–20%) and a higher percentage as DMAs (60–80%) [6]. Higher percentages of iAs and MMAs in urine may indicate poorer metabolism/biotransformation of iAs and have been linked to some of the adverse health effects including cancer, and cardiovascular disease [9]. In relation to infant health, we have shown that higher urinary MMAs and %MMAs in pregnant women are associated with adverse health outcomes in infants, specifically lower birth weight [10]. Individual iAs methylation capacities vary across individuals and depend on other numerous factors, such as dose and duration of exposure to iAs, co-exposure to other toxicants, nutritional status, gender, age, and genetic factors [4, 9, 11, 12].
The biotransformation, i.e. methylation, of arsenic is conducted by the enzyme arsenic (+3 oxidation state) methyltransferase (AS3MT) [13]. Confirming the central role of this enzyme in arsenic methylation, a dramatic decrease in arsenic methylation capacity has been observed in AS3MT knock-out mice and in HepG2 cells where it has been silenced [14, 15]. Additionally, single nucleotide polymorphisms (SNPs) in the AS3MT gene have been associated with altered arsenic methylation capabilities [16–20]. Further evidence supporting the role of AS3MT as a primary mediator of arsenic metabolism is a genome-wide association (GWAS) study in Bangladesh highlighting a strong association between arsenic metabolism and the chromosome 10q24.32 region, the genomic location of AS3MT [21]. AS3MT polymorphisms have been associated with the metabolic patterns of iAs in individuals, and these patterns have been shown to differ among racial or ethnic groups [16, 20]. AS3MT polymorphisms have also been associated with the level of AS3MT expression in peripheral blood [22].”
The aim of the present study was to examine the relationship and potential interactions between maternal and fetal genotypes for AS3MT examining seven targeted SNPs as they relate to iAs metabolism indicators and infant birth outcomes in the BEAR pregnancy cohort located in Goméz Palacio, Mexico [1, 10, 23, 24]. SNPs evaluated in this study have been previously shown to be associated with arsenic metabolism, AS3MT gene expression in blood, or iAs associated diseases [16, 18, 20–22, 25–37]. However, to date the relationships between maternal and fetal AS3MT genotypes as predictors of maternal metabolism and infant health outcomes have not been examined.
2. Materials and Methods
2.1 Ethics Statement, Study Subjects and Sample Collection
This study was approved by the Institutional Review Boards of the University of North Carolina at Chapel Hill and Universidad Juárez del Estado de Durango (UJED). A total of 200 pregnant women residing in Gómez Palacio, in the State of Durango, Mexico, were recruited at the General Hospital of Gómez Palacio to participate in the BEAR prospective pregnancy cohort. At birth, cord blood serum samples and birth outcome measures were collected from all infants born to recruited mothers. Requirements for participation, recruitment information, sampling and determination of urinary and drinking water arsenicals, and birth outcome measures have been reported previously [10, 23]. Briefly, water samples were collected from the participants’ stated main source of drinking water by a member of the research team. The concentrations of DW-iAs were measured using hydride generation-atomic absorption spectrometry (HG-AAS) system as described previously [38, 39]. The limit of detection (LOD) for DW-iAs was 0.456 mg/L. Maternal spot urine samples were collected at the time of delivery, immediately placed in a cryovial, and stored in liquid nitrogen. Samples were shipped at −80 C to the University of North Carolina at Chapel Hill (Chapel Hill, NC) for analysis. The specific gravity (SG) of each urine sample was measured using a handheld refractometer (Reichert TX 400 #13740000; Reichert Inc., Depew, NY). The major arsenical species, specifically iAs and its monomethylated and dimethylated metabolites (MMAs and DMAs), were measured using HG-AAS with cryotrapping [39, 40]. The LOD for urinary iAs, MMAs, and DMAs were 0.2 ng/mL, 0.1 ng/mL, and 0.1 ng/mL, respectively. U-tAs was defined as the SG-adjusted sum of iAs, MMAs (trivalent + pentavalent monomethylated arsenicals), and DMAs (trivalent + pentavalent dimethylated arsenicals). Each urine sample was adjusted using the following formula: (mean measured SG-1)/(individual measured SG-1) [41], where the mean SG of the cohort was 1.014 g/ml. Levels of arsenic in drinking water and urine that were below the LOD were converted to values according to the formula: LOD/(√2) [42]. We selected not to adjust by creatinine as it has been shown to be associated with arsenic methylation efficiency [12].
2.2 DNA Isolation and Genotyping
DNA was isolated from the 200 maternal whole blood and 200 fetal cord blood samples using the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA) according to the manufacturer’s protocol and stored at −80oC. Quality and concentration of DNA was evaluated on a NanoDrop 2000c UV-Vis spectrophotometer (Thermo Scientific). Seven polymorphisms in the AS3MT gene (ID: 57412; NM_020682.3; NP_065733.2) that have been previously associated with inter-individual differences in iAs metabolism and/or AS3MT expression were evaluated. These SNPs were evaluated using functionally tested (rs7085104, rs3740400, rs3740390, rs11191439, rs10748835, rs1046778) or validated (rs3740393) TaqMan assays purchased from AB Applied Biosystems (Foster City, CA). The ABI Dual 384-Well GeneAmp PCR System 9700 and ABI PRISM 7900HT Sequence Detection System from Applied Biosystems were used for genotyping and the ABI SDS software for data analysis. Here, 10% of randomly selected samples were used for a quality control assessment. In addition, 10% of samples were independently reanalyzed using the LightCycler® 480 Instrument (Roche) and genotypes evaluated by Endpoint Genotyping LC480 software.
2.3 Statistical Analysis
All statistical analyses were carried out using R [43]. SNP genotypes were numerically coded as zero (wild type homozygote, zero copies of the minor/less common allele), one (heterozygote, one copy of the minor allele) or two (variant homozygote, two copies of the minor allele). The Hardy-Weinberg equilibrium (HWE) test was performed for each SNP. Three women with missing genotype and/or birth outcome information were excluded from the analysis, bringing the final sample size to 197 mother-infant pairs (n=394 subjects). Allele frequencies were compared to two separate populations, located in San Antonio de los Cobres, Argentina and Matlab, Bangladesh, that had been previously genotyped for the seven SNPs in AS3MT [22].
Multiple linear regression was performed to examine the relationships between maternal and/or fetal alleles for each of the seven SNPs, six iAs metabolite levels/percentages and six birth outcomes/measures. The iAs metabolites investigated were U-iAs, U-MMAs, and U-DMAs, and their percentages: %iAs, %MMAs and %DMAs. The six birth outcomes/measures assessed were gestational age (wks), birthweight (g), birthweight/gestational age, placental weight (g), length (cm), and head circumference (cm). This allowed for comparisons to the primary predictors, maternal and/or fetal AS3MT genotype, while controlling for the effects of several important cofounders known to affect arsenic metabolism. A priori covariates were selected based on their known relationship to birth outcomes as well as metabolism. These factors included: U-tAs (total urinary arsenic as a measure of exposure), maternal age, smoking status, drinking status, and education level (a surrogate used for socioeconomic status). While there can be differences in iAs metabolism over pregnancy, all samples were collected at the time of birth thus minimizing metabolism differences during times of pregnancy [44, 45]. The data were analyzed for outliers and determined not to be influenced by outliers. Five models were constructed and tested. The primary model (Model I) included maternal AS3MT genotype as a predictor, controlling for fetal sex. Additionally, because there are known sex specific effects of arsenic on infant biomarkers and birth outcomes [10, 46, 47], a fetal sex-stratified model was included (Model II). To investigate potential contributions from the fetal AS3MT genotypes, three additional models were used. Model III assessed the interaction between maternal and fetal genotypes. Model IV assessed maternal and fetal genotype individually, and Model V assessed fetal genotype alone. Beta (β) coefficients were calculated representing the estimated change in iAs metabolite levels or percentages or birth outcomes/measures given the addition of one copy of the minor allele. Specifically, heterozygotes were compared to the referent group, the wild type homozygous group carrying zero copies of the minor allele. As the SNPs analyzed were in high linkage disequilibrium, for all models an adjusted p-value threshold of ≤0.01 was used for significance representing a correction for five tests (p=0.05/5) as detailed [48]. Additionally, because of this high linkage disequilibrium, a haplotype-based analysis was further performed using the R package, haplo.stats [49]. A permutation-based haplotype analysis was performed on all 21 possible SNP pairs, and on three selected three-SNP sets, with significance set at p≤0.05.
3. Results
3.1 Characteristics of the study population
Selected maternal demographic characteristics, indicators of iAs exposure, pregnancy and birth outcomes of the mother-infant pairs of BEAR cohort are described in Table 1 and reported in detail elsewhere [10]. Briefly, all 200 women participating in the study were of Hispanic origin, and with good overall health status. Their average age was 24 years, and most had singleton pregnancies. Among all births, 1.5% were preterm (<37 weeks). Male infants (52%) on average had a significantly (p = 0.0003) higher birth weight (3453 g) compared to females (3215 g). Of all infants, 2% were of low birth weight, 14% were small for gestational age, while approximately 10% were large for gestational age. The infants had an average length of 50 cm, and head circumference of 35 cm (Table 1).
Table 1.
Selected demographic characteristics and iAs exposure indicators of the BEAR study.
| Characteristic | n (%)+ | Mean, Median (Range) |
|---|---|---|
| Maternal Age at Delivery (years) | - | 24, 23 (18–41) |
| Education | ||
| Less than High School | 50 (25.1%) | - |
| High School or Above | 149 (74.5%) | - |
| Smoking Status During Pregnancy | ||
| Non-smokers | 186 (93.0%) | - |
| Current smokers | 13 (7.0%) | - |
| Alcohol Consumption During Pregnancy | ||
| None | 159 (79.5%) | - |
| Some | 41 (20.5%) | - |
| Seafood Consumption During Pregnancy | ||
| None | 155 (78.3%) | - |
| Some | 43 (21.7%) | - |
| Infant Sex | ||
| Male | 104 (52.0%) | - |
| Female | 96 (48.0%) | - |
| Birth Outcomes | ||
| Gestational Age (weeks) | ||
| All | - | 39, 40 (34–42) |
| <37 weeks (preterm) | 3 (1.5%) | - |
| >37 weeks (not preterm) | 197 (98.5%) | - |
| Birth weight (g) | ||
| All | - | 3339, 3355 (1800–5120) |
| Male | - | 3453, 3490 (2100–5120)* |
| Female | - | 3215, 3150 (1800–4200) |
| Low Birth Weight (LBW) | 4 (2.0%) | - |
| Small for Gestational Age (SGA) | 28 (14.0%) | - |
| Large for Gestational Age (LGA) | 19 (9.5%) | - |
| Placental Weight (g) | - | 648, 640 (390–1070) |
| Length (cm) | - | 50, 50 (40–59) |
| Head Circumference (cm) | - | 35, 35 (31–38) |
| Exposure measures | ||
| DW-iAs (μg/L) | - | 24.6, 13.0 (<LOD#- 236.0) |
| U-tAs (μg/L) | - | 37.5, 23.3 (4.3–319.7) |
| Metabolism of Arsenic Indicators | ||
| U-iAs (μg/L) | - | 2.1, 1.3 (<LOD#-23.0) |
| U-MMAs (μg/L) | - | 2.3, 1.4 (0.12–18.2) |
| U-DMAs (μg/L) | - | 33.1, 20.6 (1.4–292.5) |
| iAs (%) | - | 6.1, 5.3 (0.77–45.1) |
| MMAs (%) | - | 6.4, 6.0 (0.68–24.9) |
| DMAs (%) | - | 87.6, 88.5 (32.7–96.7) |
Differences in n based on missing demographic data.
Significant difference in means (p=0.0003) between males and females.
Limit of detection (LOD) for DW-iAs=0.46 μg/L
Limit of detection (LOD) for U-iAs=0.2 μg/L
Limit of detection (LOD) for U-MMAs=0.1 μg/L
Limit of detection (LOD) for U-DMAs=0.1 μg/L
Indicators of iAs exposure included the concentration of iAs in drinking water (μg/L; DW-iAs) and maternal concentrations of iAs and its methylated metabolites measured in urine, MMAs and DMAs (μg/L; U-iAs, U-MMAs, and U-DMAs). Due to the instability of trivalent methylated arsenicals [37], only a sum of trivalent (AsIII) and pentavalent (AsV) arsenicals was measured. The levels of iAs in drinking water samples of the cohort ranged from the LOD of 0.46 μg/L up to 236 μg/L. Of these drinking water samples, 107 (53%) had DW-iAs that exceeded the WHO standard (10 μg/L) and 56 (28%) exceeded Mexico’s standard of 25 μg/L. The mean and median DW-iAs of the cohort were 24.6 μg/L and 13.0 μg/L, respectively (Table 1). As with DW-iAs, a range of U-iAs, U-MMAs and U-DMAs was observed (Table 1). Most of the urine samples (95%) had detectable levels of U-iAs and U-MMAs with ranges of <LOD (0.2 μg/L)-23.0 μg/L and < 0.12 μg/L-18.2 μg/L, respectively. DMAs were detected in all urine samples and ranged from 1.4–292.5 μg/L. Total maternal urinary arsenic (U-tAs), defined as the sum of U-iAs, U-MMAs, and U-DMAs, ranged from 4.30 μg/L to 319.7 μg/L. The average percents of the iAs metabolites with regard to U-tAs were 6.1% iAs, 6.4% MMAs, and 87.6% DMAs, which is in agreement with studies that report a generally higher methylation capacity of pregnant women as compared to non-pregnant women [6]. For this cohort, we have previously reported a significant positive correlation between DW-iAs and U-tAs (r = 0.51; p<0.0001) [10].
3.2 Allele frequency assessment
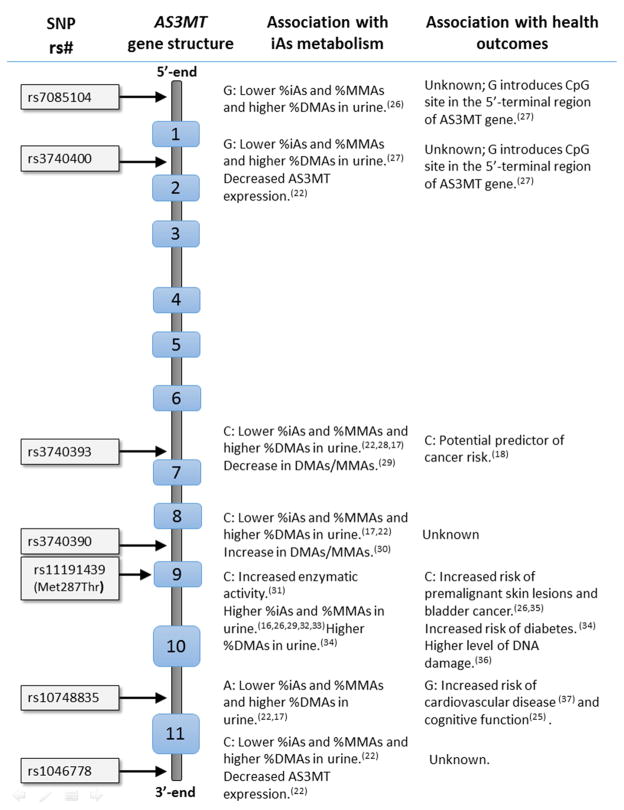
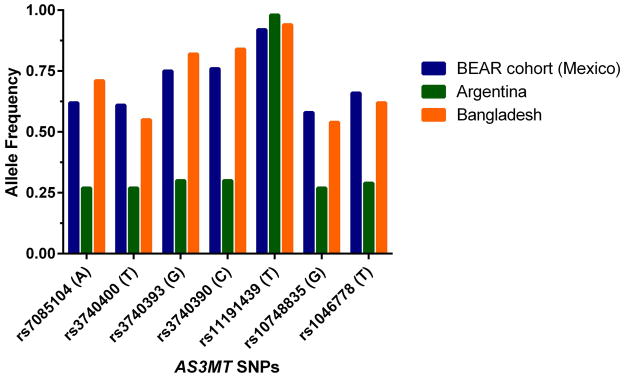
The women and infants of the BEAR cohort were classified into one of three categories depending upon their genotypes as wild-type homozygotes (i.e., carriers of two copies of the major allele), variant homozygotes (carriers of two copies of the minor allele) or heterozygotes (carriers of one copy of the major allele and one copy of the minor allele). Six of the tested SNPs are located within intronic regions (rs7085104, rs3740400, rs3740393, rs3740390, rs10748835 and rs1046778) of AS3MT. One non-synonymous SNP present in exon nine alters the amino acid sequence of the AS3MT protein (rs11191439, Met287Thr). The SNP positions within the AS3MT gene and previously published associations between alleles within SNPs, iAs metabolism indicators, AS3MT expression, and health outcomes are summarized in Figure 1. A comparison of the allele frequencies for the BEAR cohort relative to genotype frequencies found in two independent cohorts, one in San Antonio de los Cobres, Argentinea (n=176) and one in Matlab, Bangladesh (n=359) are provided (Figure 2, Table 2) [22]. These data show that the allele frequencies of women in Goméz Palcio are generally concordant with those in Matlab, Bangladesh, but differ from those in San Antonio de los Cobres, Argentina. All examined SNPs passed the Hardy-Weinberg equilibrium test at p<0.05. Five pairs of SNPs, rs7085104 and rs3740400, rs3740400 and rs10748835, rs3740393 and rs3740390, rs7085104 and rs10748835, rs10748835 and rs1046778, were in strong linkage disequilibrium with r2 = 0.95, 0.75, 0.74, 0.72, and 0.71, respectively (Figure 3).
Figure 1.
Position of single nucleotide polymorphisms (SNPs) within the AS3MT gene and known associations with iAs biotransformation and health outcomes. Arrows indicate the region of the SNP locations within the gene. Light blue rectangles represent the 11 exonic regions of the gene.
Figure 2.
Allelic frequency comparison of the Goméz Palacio, Mexico cohort to populations in San Antonio de los Cobres, Argentinea (n=176) or Matlab, Bangladesh (n=359) as detailed in [22].
Table 2.
Allele frequencies and Hardy-Weinberg p-values for the BEAR cohort.
| SNP rs | Genotype | % (N) | Allele Frequency BEAR cohort | Hardy- Weinberg p-value | Allele Frequency Argentina* | Allele Frequency Bangladesh* |
|---|---|---|---|---|---|---|
| rs7085104 (A>G) | AA | 38.5 (76) | ||||
| AG | 45.6 (90) | A allele: 0.62 | A allele: 0.27 | A allele: 0.71 | ||
| GG | 15.2 (30) | G allele: 0.38 | 0.78 | G allele: 0.73 | G allele: 0.29 | |
| ND | 0.5 (1) | |||||
| rs3740400 (T>G) | TT | 37.4 (73) | ||||
| TG | 46.7 (92) | |||||
| GG | 15.7 (31) | T allele: 0.61 | T allele: 0.27 | T allele: 0.55 | ||
| ND | 0.5(1) | G allele: 0.39 | 0.91 | G allele: 0.73 | G allele: 0.45 | |
| rs3740393 (G>C) | GG | 58.9 (116) | ||||
| GC | 32.0 (63) | G allele: 0.75 | G allele: 0.30 | G allele: 0.82 | ||
| CC | 9.1 (18) | C allele: 0.25 | 0.05 | C allele: 0.70 | C allele: 0.18 | |
| ND | 0 (0) | |||||
| rs3740390 (C>T) | CC | 58.4 (115) | ||||
| CT | 34.5 (68) | C allele: 0.76 | C allele: 0.30 | C allele: 0.84 | ||
| TT | 6.1 (12) | T allele: 0.24 | 0.76 | T allele: 0.70 | T allele: 0.16 | |
| ND | 1.0 (2) | |||||
| rs11191439 (T>C) | TT | 83.8 (165) | ||||
| (Met287Thr) | TC | 14.7 (29) | T allele: 0.92 | T allele: 0.98 | T allele: 0.94 | |
| CC | 0.5 (1) | C allele: 0.08 | 0.81 | C allele: 0.02 | C allele: 0.06 | |
| ND | 1.0 (2) | |||||
| rs10748835 (G>A) | GG | 31.5 (62) | ||||
| GA | 52.8 (104) | G allele: 0.58 | G allele: 0.27 | G allele: 0.54 | ||
| AA | 15.2 (30) | A allele: 0.42 | 0.25 | A allele: 0.73 | A allele: 0.46 | |
| ND | 0.5 (1) | |||||
| rs1046778 (T>C) | TT | 44.2 (87) | ||||
| TC | 43.1 (85) | T allele: 0.66 | T allele: 0.29 | T allele: 0.62 | ||
| CC | 12.2 (24) | C allele: 0.34 | 0.74 | C allele: 0.71 | C allele: 0.38 | |
| ND | 0.5 (1) |
N – Number of subjects;
Allele frequencies from Engstrom. K., Vahter, M., et al. (2011) EHP 119:182-188.
Figure 3.
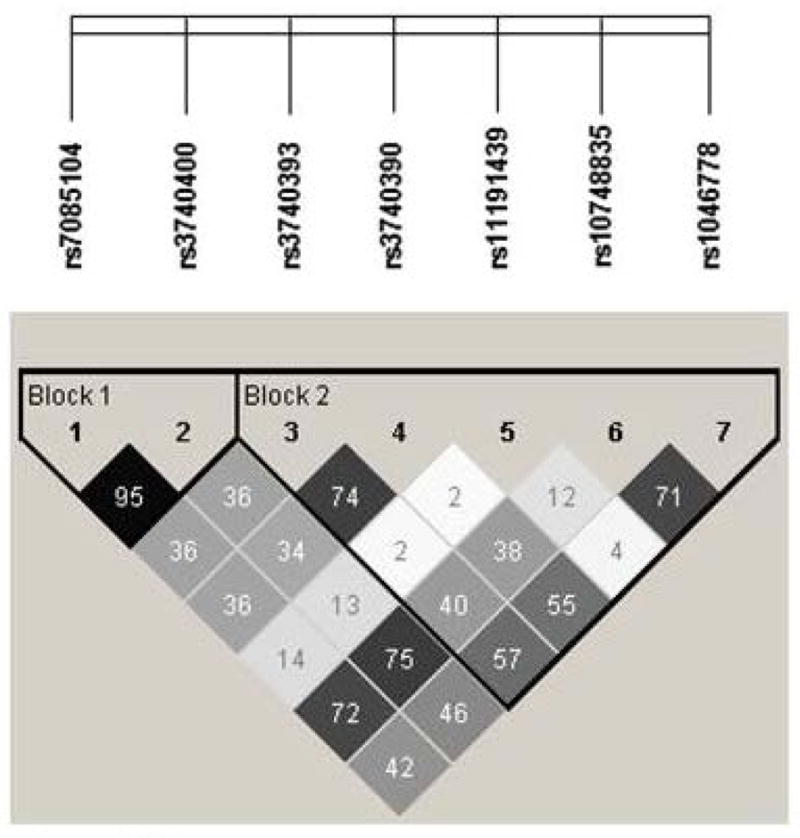
Linkage disequilibrium values (R-squared) for seven AS3MT polymorphisms in the Goméz Palacio, Mexico cohort.
3.3.1 Association between maternal AS3MT genotype and arsenic metabolites
For all alleles of each of the seven SNPs analyzed, multiple linear regression analyses were conducted using the models specified above. Associations were examined between maternal AS3MT alleles and urinary iAs metabolite levels and percentages (Table 3, Supplemental Table 1). Alleles of five of the seven SNPs (rs7085104, rs3740400, rs3740393, rs3740390, and rs1046778) showed significant (p<0.01) associations with U-MMAs, U-DMAs, %MMAs, or %DMAs while rs11191439 showed no significance with iAs or any of the urinary metabolites (Table 3, Supplemental Table 1). Alleles of SNP rs10748835 showed marginally significant (p<0.05) associations with U-MMAs and %MMAs (Table 3). As indicated by the beta (β) coefficients presented in Table 3, lower U-MMAs and %MMAs and higher U-DMAs and %DMAs were associated with the presence of the minor/less common allele for all alleles of these SNPs (Table 3). This general trend supports that the major maternal AS3MT alleles are associated with higher %MMAs and lower %DMAs in urine.
Table 3.
Association between maternal alleles of AS3MT single nucleotide polymorphisms (SNPs) and urinary iAs metabolite levels and percentages.
| SNP rs (major allele>minor allele) | U-iAs (βc) | U-MMAs (β) | U-DMAs (β) | %iAs (β) | %MMAs (β) | %DMAs (β) |
|---|---|---|---|---|---|---|
| Model Ia | ||||||
| rs7085104 (A>G) | - | −0.453++ (−0.748, −0.157) | 0.768++ (0.214, 1.321) | - | −1.050++ (−1.645, −0.454) | 1.729++ (0.492, 2.966) |
| rs3740400 (T>G) | - | −0.452++ (−0.156, −0.747) | 0.731++ (1.286, 0.176) | - | −1.028++ (−0.431, −1.625) | 1.613+ (2.854, 0.372) |
| rs3740393 (G>C) | - | −0.422++ (−0.109, −0.736) | 0.669+ (1.257, 0.08) | - | −1.182++ (−0.555, −1.81) | 1.705+ (3.017, 0.394) |
| rs3740390 (C>T) | - | −0.494++ (−0.833, −0.154) | 0.703+ (0.061, 1.345) | - | −1.253++ (−1.94, −0.565) | 1.833+ (0.396, 3.269) |
| rs11191439 (T>C) d | - | - | - | - | - | - |
| rs10748835 (G>A) | - | −0.324+ (−0.637, −0.012) | - | - | −0.815+ (−1.446, −0.183) | - |
| rs1046778 (T>C) | - | −0.40++ (−0.707, −0.098) | 0.581+ (0.008, 1.153) | - | −1.012++ (−1.625, −0.399) | 1.510+ (0.235, 2.785) |
| Model II: Malesb | ||||||
| rs7085104 (A>G) | −0.390++ (−0.666, −0.115) | −0.356+ (−0.661, −0.052) | 0.731++ (0.236, 1.226) | −0.809+ (−1.522, − 0.096) | −1.091++ (−1.751, −0.432) | 1.814++ (0.711, 2.918) |
| rs3740400(T>G) | −0.313+ (−0.035,− 0.59) | −0.370+ (−0.069, −0.671) | 0.665++ (1.160, 0.170) | - | −1.155++ (−0.506, −1.805) | 1.658++ (2.763, 0.552) |
| rs3740393 (G>C) | −0.410++ (−0.109, −0.71) | −0.464++ (−0.138, −0.791) | 0.858++ (1.393, 0.324) | −1.005++ (0.235, 1.776) | −1.323++ (−0.613, −2.033) | 2.201++ (3.390, 1.011) |
| rs3740390 (C>T) | −0.386+ (−0.712, −0.06) | −0.519++ (−0.869, −0.169) | 0.899++ (0.324, 1.475) | −0.885+ (−1.721, − 0.049) | −1.401++ (−2.163, −0.639) | 2.366++ (1.095, 3.638) |
| rs11191439 (T>C) d | - | 1.064++ (0.480, 1.648) | −1.585++ (−2.562, −0.607) | - | 1.362+ (0.002, 2.721) | - |
| rs10748835 (G>A) | - | - | - | - | −0.829+ (−1.542, −0.117) | 1.364+ (0.171, 2.557) |
| rs1046778 (T>C) | - | −0.425++ (−0.727, −0.123) | 0.675++ (0.174, 1.175) | - | −1.103++ (−1.765, −0.441) | 1.807++ (0.698, 2.917) |
| Model II: Femalesb | ||||||
| rs7085104 (A>G) | - | - | - | - | - | - |
| rs3740400(T>G) | - | - | - | - | - | - |
| rs3740393 (G>C) | - | - | - | - | - | - |
| rs3740390 (C>T) | - | - | - | - | - | - |
| rs11191439 (T>C) d | - | - | - | - | - | - |
| rs10748835 (G>A) | - | - | - | - | - | - |
| rs1046778 (T>C) | - | - | - | - | - | - |
p<0.05
p<0.01 (adjusted p-value).
Model I included maternal genotype as a primary predictor, controlling for the following covariates: U-tAs, maternal age, smoking status, drinking status, education level and fetal sex.
Model II included maternal genotype as a primary predictor, controlling for the following covariates: U-tAs, maternal age, smoking status, drinking status, and education level, stratified by fetal sex.
Beta (β) coefficients represent the estimated change in iAs metabolite levels or percentages resulting from the addition of one copy of the minor allele.
rs11191439 is a non-synonymous SNP in exon nine known to alter the amino acid sequence of AS3MT (Met287Thr).
Associations between maternal alleles in AS3MT SNPs and maternal urinary arsenic metabolite levels and percentages were further stratified based upon the sex of the infant (Model II) (Table 3). Generally, associations between maternal AS3MT alleles and levels and %MMAs and %DMAs were significant if women were pregnant with a male child, but this was not the case for women pregnant with a female child. The presence of the minor allele in rs1191439 (Met287Thr) in women pregnant with a male, but not female, child was associated with increased U-MMAs and %MMAs and decreased U-DMAs and %DMAs. Interestingly, this is the opposite relationship as was observed for the intronic SNPs, rs7085104, rs3740400, rs3740393, rs3740390, and rs1046778, where the minor allele was associated with decreased U-MMAs and %MMAs and increased U-DMAs and %DMAs. Taken together, significant negative associations were observed between the minor alleles and %MMAs and positive associations between the minor alleles and %DMAs for all tested genotypes, with this relationship inverted for alleles of SNP rs11191439. No significant associations were found between maternal SNPs and urinary arsenic metabolite levels when women were pregnant with a female child (Table 3, Supplemental Table 1). The stratified analyses highlight a strong relationship between maternal genotype and fetal sex, with significant associations between maternal alleles of AS3MT SNPs, urinary arsenic metabolites, and birth outcomes in women pregnant with males.
3.3.2 Association between maternal AS3MT genotype and birth outcome/measures
Maternal alleles of AS3MT SNPs were also compared to six birth outcomes/measures (Table 4, Supplemental Table 2). Statistical significance was observed between alleles of rs3740393 and placental weight (β=40.873, p=0.0056). Alleles of this same SNP showed a marginally significant (p=0.02) relationship to birthweight. The majority of relationships observed between maternal alleles of AS3MT and birth outcomes/measures were only marginally significant (p≤0.05). Overall, fewer significant associations were seen between maternal alleles of AS3MT and birth outcomes than between alleles and maternal urinary arsenic metabolite levels. Notably, for the majority of the alleles tested, there were positive associations between the minor alleles and birth outcomes/measures.
Table 4.
Association between maternal alleles of AS3MT single nucleotide polymorphisms (SNPs) and birth outcomes and measures.
| SNP rs (major allele>minor allele) | Gestational Age (wks) (βc) | Birth Weight (g) (β) | BW/GA (β) | Placental Weight (g) (β) | Length (cm) (β) | Head Cir. (cm) (β) |
|---|---|---|---|---|---|---|
| Model Ia | ||||||
| rs7085104 (A>G) | - | - | - | - | - | - |
| rs3740400(T>G) | - | - | - | - | 0.565+ (1.107, 0.023) | - |
| rs3740393 (G>C) | - | 113.521+ (210.685, 16.357) | - | 40.873++ (69.651, 12.095) | - | - |
| rs3740390 (C>T) | - | 121.003+ (15.253, 226.754) | 2.896+ (0.245, 5.548) | - | - | - |
| rs11191439 (T>C) d | - | - | - | - | - | - |
| rs10748835 (G>A) | - | - | - | - | - | - |
| rs1046778 (T>C) | - | 102.812+ (8.861, 196.764) | - | - | - | - |
| Model II: Malesb | ||||||
| rs7085104 (A>G) | - | - | - | - | - | - |
| rs3740400(T>G) | - | - | - | - | - | - |
| rs3740393 (G>C) | 0.331+ (0.731, −0.035) | - | - | - | - | - |
| rs3740390 (C>T) | - | - | - | - | - | - |
| rs11191439 (T>C) d | - | - | - | - | - | - |
| rs10748835 (G>A) | 0.414+ (0.057, 0.747) | - | - | - | 0.865+ (−0.161, 1.709) | - |
| rs1046778 (T>C) | 0.341+ (−0.071, 0.631) | - | - | - | - | - |
| Model II: Femalesb | ||||||
| rs7085104 (A>G) | - | - | - | - | - | - |
| rs3740400(T>G) | - | - | - | - | - | - |
| rs3740393 (G>C) | - | - | - | 58.405++ (97.726, − 2.951) | - | - |
| rs3740390 (C>T) | - | - | - | 54.039+ (−20.569, 91.037) | - | - |
| rs11191439 (T>C) d | - | - | - | - | - | - |
| rs10748835 (G>A) | - | - | - | - | - | - |
| rs1046778 (T>C) | - | - | - | - | - | - |
p<0.05
p<0.01 (adjusted p-value).
Model I included maternal genotype as a primary predictor, controlling for the following covariates: U-tAs, maternal age, smoking status, drinking status, education level and fetal sex.
Model II included maternal genotype as a primary predictor, controlling for the following covariates: U-tAs, maternal age, smoking status, drinking status, and education level, stratified by fetal sex.
Beta (β) coefficients represent the estimated change in birth outcome/measures resulting from the addition of one copy of the minor allele.
rs11191439 is a non-synonymous SNP in exon nine known to alter the amino acid sequence of AS3MT (Met287Thr).
Subsequently, associations between AS3MT alleles and birth outcomes/measures were stratified based upon the sex of the infant (Model II) (Table 4, Supplemental Table 2). Several birth outcomes were marginally significant (p≤0.05) in relationship to AS3MT genotype when stratified by fetal sex. Alleles of three SNPs (rs3740393, rs10748835 and rs1046778) were marginally significant (p≤0.05) and positively associated with gestational age among women pregnant with male infants. Additionally, women pregnant with male infants showed a marginally significant (p≤0.05) positive association between alleles of rs10748835 and length at birth (Table 4). In the case of women pregnant with female infants, alleles of rs3740390 and rs3740393 were positively associated with placental weight (Table 4, Supplemental Table 2).
3.4.1 Association between maternal and fetal AS3MT genotypes and arsenic metabolites
Based on the sex-specific nature of the relationship between maternal AS3MT genotype and maternal metabolism, we set out to investigate whether fetal genotype might influence maternal iAs metabolism. First, a full model with main effects of both maternal and fetal AS3MT genotypes and their interactions was considered (Model III, Supplemental Table 1). Interestingly, most of the interactions between maternal and fetal genotypes were insignificant in their relationship to maternal iAs metabolism. A marginally significant (p=0.0188) maternal-fetal genotype interaction was observed for SNP rs10748835 with %MMAs. The full model was subsequently reduced by removing the interaction term to further determine the contribution of fetal genotype (Model IV). Only the maternal AS3MT alleles showed statistical significance in association with maternal iAs metabolism indicators (Supplemental Table 1). Finally, the impact of fetal AS3MT genotype alone was tested with no significant associations found with iAs metabolism indicators (Model V) (Supplemental Table 1).
3.4.2 Association between maternal and fetal AS3MT genotypes and birth outcome/measures
We next investigated whether fetal AS3MT genotype is associated with infant outcomes/measures using the same models described above. As observed in the tests for association with maternal iAs metabolism indicators, there were no statistically significant results when testing the interaction between maternal and fetal genotypes (Supplemental Table 2). The reduced model used to examine the maternal and fetal AS3MT genotypes separately (Model IV) showed that only the maternal AS3MT genotypes showed statistical significance in association with infant outcomes/measures. Fetal alleles for rs3740390 and rs1046778 showed marginal significance (p=0.0157 and 0.0381, respectively) with placental weight. Finally, when considering the impact of fetal genotype for AS3MT alone (Model V), there were two significant associations between placental weight and fetal alleles of rs3740390 (p=0.0024) and rs1046778 (p=0.0097). Additionally, alleles for two fetal SNPs, rs37430400 and rs3740393 showed marginally significant associations with placental weight (p=0.0500 and 0.0157, respectively).
3.5 Association of maternal AS3MT haplotypes with urinary arsenicals and birth outcomes
Haplotype analysis was also conducted on all possible SNP-pairs and three selected three-SNP sets. The comparison of paired haplotypes showed results similar to the analysis of the alleles of individual SNPs. The majority of associations for pairwise haplotypes were found between maternal U-MMAs and %MMAs, with 19 out of 21 pairwise comparisons showing significant association with U-MMAs, and all 21 pairwise comparisons associated with %MMAs (Supplemental Table 3). Typically, haplotypes where the SNP pair consisted of both major alleles were associated with increased U-MMAs and %MMAs, while haplotypes where the SNP pair consisted of both minor alleles were associated with decreased U-MMAs and %MMAs, indicating the additivity of the SNPs. This is also confirmed by the observation that haplotypes where the SNP pair consisted of one major allele and one minor allele typically resulted in non-significant results. In relation to DMAs, seven of the pairwise comparisons were associated with U-DMAs and 10 of the pairwise comparisons were associated with %DMAs (Supplemental Table 3). Specifically, major allele haplotypes were associated with decreased U-DMAs and %DMAs, minor allele haplotypes were associated with increased U-DMAs and %DMAs and heterozygote allele haplotypes were typically nonsignificant. There were no associations found between U-iAs or %iAs for any of the pairwise haplotype analyses. While there were fewer associations between pairwise haplotypes and birth outcomes, a total of six of the pairwise haplotypes displayed an association with either birthweight, birthweight/gestational age, or placental weight (Supplemental Table 3).
The three-SNP cluster haplotypes were determined based upon their linkage disequilibrium coefficients (Figure 3). All three haplotypes were associated with %MMAs. Specifically, major allele haplotypes were associated with elevated %MMAs, and minor allele haplotypes were associated with reduced %MMAs (Supplemental Table 4). There were two significant associations between placental weight and birthweight/gestational age and the cluster consisting of rs7085104/rs3740400/rs1046778. No other cluster displayed a significant relationship with any of the birth outcomes/measures (Supplemental Table 4).
4. Discussion
We have recently demonstrated that iAs metabolism in pregnant women is associated with differential health outcomes in infants, where elevated U-MMAs ad %MMAs in maternal urine were associated with lower birth weight [10]. To examine whether this metabolism and subsequent birth outcomes is influenced by maternal and/or fetal genotype, we focused on polymorphisms within the primary gene associated with iAs metabolism, namely AS3MT [13, 14]. Such nucleotide differences have been shown to alter the efficiency of the iAs metabolism as measured in urinary metabolite profiles [22, 27]. Seven SNPs in AS3MT were targeted for analysis because of their known associations with iAs metabolism, AS3MT expression and/or diseases [16, 18, 20–22, 25–37]. The results demonstrate that maternal genotype for AS3MT is associated both with maternal metabolism iAs and to infant birth outcomes/measures, albeit to a lesser extent. Interestingly, we also demonstrate that the association between maternal AS3MT genotype and maternal iAs metabolism profiles was strong when women were pregnant with a male fetus, a finding that was independent of fetal genotype for AS3MT. This suggests that the sex-specific effects that were observed are not driven by fetal genotype of AS3MT. To our knowledge this is the first study to simultaneously examine the relationship of maternal and fetal AS3MT genotype with urinary arsenic metabolites in pregnant women and infant health outcomes.
We demonstrate that alleles for five of the seven SNPs (rs7085104, rs3740400, rs3740393, rs3740390, and rs1046778) were associated with iAs metabolism indicators. In the present study we observed that the minor alleles for these SNPs were associated with lower %MMAs and higher %DMAs. In contrast, the major or more common alleles were associated with higher %MMAs and lower % DMAs suggesting that the majority of individuals in the cohort are at greater risk for an elevated urinary MMA phenotype. This finding is of concern given the known links between elevated %MMAs and detrimental health outcomes including premalignant skin lesions, DNA damage in children and increased risk for bladder cancer [26, 35, 36]. Additionally, elevated levels and proportions of MMAs, have been associated with poorer fetal health outcomes, specifically lowered birth weight in this same cohort [10].
The observation that the minority of subjects carry alleles for a lower urinary MMA phenotype is similar to previous studies that have shown that the minor alleles for six of the SNPs (rs7085104, rs3740400, rs3740393, rs3740390, rs10748835 and rs1046778) evaluated are associated with lower %iAs and %MMAs and higher %DMAs in urine [11, 17, 22, 26–34, 50]. The only currently known exception is the Andean population in San Antonio de los Cobres, Argentina, where the majority of the subjects have been shown to carry AS3MT alleles associated with a reduced urinary MMA phenotype [20, 22, 27, 51]. The data from the present study highlight that the individuals with the minor alleles for rs7085104, rs3740400, rs3740393, rs3740390, rs10748835, and rs1046778, representing only 8–39% of women in total, have lower %MMAs in urine. In contrast to the findings for the alleles of the majority of the SNPs tested which showed an association between the minor allele and lower U-MMAs and %MMAs and higher U-DMAs and %DMAs, the minor or less common allele for the Met287Thr polymorphism was associated with higher levels of U-MMAs and %MMAs and lower levels of U-DMAs and %DMAs. In the present study, alleles of four of the SNPs including rs3740400, rs3740393, rs3740390, and rs1046778 were also nominally associated with birth outcomes including birth weight, placental weight, or length at birth. Here the relationships between the minor alleles and the birth outcomes were generally positively associated (i.e., increased birth weight, increased placental weight, increased length at birth) along with the previously demonstrated lower proportions of MMAs and higher proportions of DMAs. The results from the present study suggest that a pregnant woman’s ability to metabolize iAs is dependent upon her AS3MT genotype and this metabolism influences the health of her unborn child.
Interestingly, fetal sex-stratified analysis revealed stronger associations between maternal genotype for AS3MT and maternal metabolites when women were pregnant with a male fetus. As determined through fetal AS3MT genotype testing, this effect was not related to the fetal genotype of AS3MT. After stratifying for the sex of the infant, alleles for six of the SNPs were significantly associated with metabolite levels in mothers pregnant with male fetuses, in strong contrast to no alleles showing association with iAs metabolite levels in mothers pregnant with female fetuses. As with the overall analysis, for women carrying male fetuses, the minor AS3MT alleles were associated with lower %MMAs and higher %DMAs, while women carrying female fetuses showed no significant associations between maternal genotype and maternal iAs metabolites. Sex-specific differences have been associated with arsenic exposure. For example, early-life exposure to arsenic has been shown to increase the incidence of cancer, cardiovascular disease, and pulmonary outcomes in sex-specific manner [52–54]. Taken together, this finding suggests that mothers may metabolize arsenic differently not only based upon their genotype for AS3MT but also depending upon whether they are pregnant with a female or male fetus. While further research is needed to determine a mechanism that may underlie this effect, the influence of sex hormones or involvement of certain sex-specific signaling pathways are likely to underlie these effects. These observations from the present study support a growing body of literature highlighting sex-specific differences in response to prenatal arsenic exposure.
While this study demonstrates novel findings of associations between maternal genotypes for AS3MT, iAs metabolism indicators and birth outcomes/measures, it is not without limitations. In the present study we did not directly control for seafood consumption which can influence U-DMA levels [55]. However, previous analyses within this cohort showed that seafood consumption did not significantly affect the association between iAs metabolite levels and birth outcomes [10]. Additionally, while we controlled for factors known to influence iAs exposure in our analysis, we were unable to control for differences in distinct nutritional status/habits of specific population, particularly nutrients known to affect iAs metabolism.
This study is the first to report significant associations between maternal alleles of AS3MT genotype and sex-specific effects on maternal iAs metabolites and infant birth outcomes. Although this study does not provide a precise molecular mechanism underlying these effects on infant birth outcomes, it supports that specific maternal metabolic profiles of arsenic depend on AS3MT genotype and that these are influenced by the sex, but not the AS3MT genotype, of the fetus. Further studies are needed to evaluate the potential long-lasting effects of prenatal iAs exposure on children’s health and determine how to best predict and protect those at increased risk for detrimental health effects.
Supplementary Material
Highlights.
Maternal genotype for AS3MT is linked to maternal metabolism iAs and to infant birth outcomes/measures.
Associations between maternal AS3MT genotype and maternal iAs metabolism profiles was strong when women were pregnant with a male fetus.
Fetal genotype was not linked to changes in maternal iAs metabolism or infant birth outcomes/measures.
The minor/less common allele was associated with decreased U-MMAs/%MMAs and increased U-DMAs/%DMAS.
Acknowledgments
Funding: This research was funded by grants from the National Institutes of Health (R01-ES109315, P42-ES007126 and T32-ES007018).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bailey KA, Laine J, Rager JE, Sebastian E, Olshan A, Smeester L, et al. Prenatal arsenic exposure and shifts in the newborn proteome: interindividual differences in tumor necrosis factor (TNF)-responsive signaling. Toxicological sciences : an official journal of the Society of Toxicology. 2014;139:328–37. doi: 10.1093/toxsci/kfu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Concha G, Vogler G, Lezcano D, Nermell B, Vahter M. Exposure to inorganic arsenic metabolites during early human development. Toxicological sciences : an official journal of the Society of Toxicology. 1998;44:185–90. doi: 10.1006/toxs.1998.2486. [DOI] [PubMed] [Google Scholar]
- 3.Hall M, Gamble M, Slavkovich V, Liu X, Levy D, Cheng Z, et al. Determinants of arsenic metabolism: blood arsenic metabolites, plasma folate, cobalamin, and homocysteine concentrations in maternal-newborn pairs. Environmental health perspectives. 2007;115:1503–9. doi: 10.1289/ehp.9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vahter M. Effects of arsenic on maternal and fetal health. Annual review of nutrition. 2009;29:381–99. doi: 10.1146/annurev-nutr-080508-141102. [DOI] [PubMed] [Google Scholar]
- 5.Smith AH, Marshall G, Liaw J, Yuan Y, Ferreccio C, Steinmaus C. Mortality in young adults following in utero and childhood exposure to arsenic in drinking water. Environmental health perspectives. 2012;120:1527–31. doi: 10.1289/ehp.1104867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vahter M. Mechanisms of arsenic biotransformation. Toxicology. 2002;181–182:211–7. doi: 10.1016/s0300-483x(02)00285-8. [DOI] [PubMed] [Google Scholar]
- 7.Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicology and applied pharmacology. 2001;176:127–44. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- 8.Styblo M, Del Razo LM, Vega L, Germolec DR, LeCluyse EL, Hamilton GA, et al. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Archives of toxicology. 2000;74:289–99. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 9.Kapaj S, Peterson H, Liber K, Bhattacharya P. Human health effects from chronic arsenic poisoning--a review. Journal of environmental science and health Part A, Toxic/hazardous substances & environmental engineering. 2006;41:2399–428. doi: 10.1080/10934520600873571. [DOI] [PubMed] [Google Scholar]
- 10.Laine JE, Bailey KA, Rubio-Andrade M, Olshan AF, Smeester L, Drobna Z, et al. Maternal arsenic exposure, arsenic methylation efficiency, and birth outcomes in the Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Mexico. Environmental health perspectives. 2015;123:186–92. doi: 10.1289/ehp.1307476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindberg AL, Ekstrom EC, Nermell B, Rahman M, Lonnerdal B, Persson LA, et al. Gender and age differences in the metabolism of inorganic arsenic in a highly exposed population in Bangladesh. Environmental research. 2008;106:110–20. doi: 10.1016/j.envres.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Basu A, Mitra S, Chung J, Guha Mazumder DN, Ghosh N, Kalman D, et al. Creatinine, diet, micronutrients, and arsenic methylation in West Bengal, India. Environmental health perspectives. 2011;119:1308–13. doi: 10.1289/ehp.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, et al. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. The Journal of biological chemistry. 2002;277:10795–803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- 14.Drobna Z, Naranmandura H, Kubachka KM, Edwards BC, Herbin-Davis K, Styblo M, et al. Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chemical research in toxicology. 2009;22:1713–20. doi: 10.1021/tx900179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drobna Z, Xing W, Thomas DJ, Styblo M. shRNA silencing of AS3MT expression minimizes arsenic methylation capacity of HepG2 cells. Chemical research in toxicology. 2006;19:894–8. doi: 10.1021/tx060076u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg AL, Kumar R, Goessler W, Thirumaran R, Gurzau E, Koppova K, et al. Metabolism of low-dose inorganic arsenic in a central European population: influence of sex and genetic polymorphisms. Environmental health perspectives. 2007;115:1081–6. doi: 10.1289/ehp.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlawicke-Engstrom K, Nermell B, Concha G, Stromberg U, Vahter M, Broberg K. Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutation research. 2009;667:4–14. doi: 10.1016/j.mrfmmm.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Chung CJ, Hsueh YM, Bai CH, Huang YK, Huang YL, Yang MH, et al. Polymorphisms in arsenic metabolism genes, urinary arsenic methylation profile and cancer. Cancer causes & control : CCC. 2009;20:1653–61. doi: 10.1007/s10552-009-9413-0. [DOI] [PubMed] [Google Scholar]
- 19.Antonelli R, Shao K, Thomas DJ, Sams R, 2nd, Cowden J. AS3MT, GSTO, and PNP polymorphisms: impact on arsenic methylation and implications for disease susceptibility. Environmental research. 2014;132:156–67. doi: 10.1016/j.envres.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Schlawicke-Engstrom K, Broberg K, Concha G, Nermell B, Warholm M, Vahter M. Genetic polymorphisms influencing arsenic metabolism: evidence from Argentina. Environmental health perspectives. 2007;115:599–605. doi: 10.1289/ehp.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, et al. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS genetics. 2012;8:e1002522. doi: 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engstrom K, Vahter M, Mlakar SJ, Concha G, Nermell B, Raqib R, et al. Polymorphisms in arsenic(+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environmental health perspectives. 2011;119:182–8. doi: 10.1289/ehp.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rager JE, Bailey KA, Smeester L, Miller SK, Parker JS, Laine JE, et al. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environmental and molecular mutagenesis. 2014;55:196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojas D, Rager JE, Smeester L, Bailey KA, Drobna Z, Rubio-Andrade M, et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicological sciences : an official journal of the Society of Toxicology. 2015;143:97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards M, Hall J, Gong G, O'Bryant SE. Arsenic exposure, AS3MT polymorphism, and neuropsychological functioning among rural dwelling adults and elders: a cross-sectional study. Environmental health : a global access science source. 2014;13:15. doi: 10.1186/1476-069X-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela OL, Drobna Z, Hernandez-Castellanos E, Sanchez-Pena LC, Garcia-Vargas GG, Borja-Aburto VH, et al. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicology and applied pharmacology. 2009;239:200–7. doi: 10.1016/j.taap.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engstrom KS, Hossain MB, Lauss M, Ahmed S, Raqib R, Vahter M, et al. Efficient arsenic metabolism--the AS3MT haplotype is associated with DNA methylation and expression of multiple genes around AS3MT. PloS one. 2013;8:e53732. doi: 10.1371/journal.pone.0053732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meza MM, Yu L, Rodriguez YY, Guild M, Thompson D, Gandolfi AJ, et al. Developmentally restricted genetic determinants of human arsenic metabolism: association between urinary methylated arsenic and CYT19 polymorphisms in children. Environmental health perspectives. 2005;113:775–81. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agusa T, Iwata H, Fujihara J, Kunito T, Takeshita H, Minh TB, et al. Genetic polymorphisms in AS3MT and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicology and applied pharmacology. 2009;236:131–41. doi: 10.1016/j.taap.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 30.Gomez-Rubio P, Meza-Montenegro MM, Cantu-Soto E, Klimecki WT. Genetic association between intronic variants in AS3MT and arsenic methylation efficiency is focused on a large linkage disequilibrium cluster in chromosome 10. Journal of applied toxicology : JAT. 2010;30:260–70. doi: 10.1002/jat.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, et al. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. The Journal of biological chemistry. 2006;281:7364–73. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez A, Xamena N, Surralles J, Sekaran C, Tokunaga H, Quinteros D, et al. Role of the Met(287)Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutation research. 2008;637:80–92. doi: 10.1016/j.mrfmmm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez A, Xamena N, Sekaran C, Tokunaga H, Sampayo-Reyes A, Quinteros D, et al. High arsenic metabolic efficiency in AS3MT287Thr allele carriers. Pharmacogenetics and genomics. 2008;18:349–55. doi: 10.1097/FPC.0b013e3282f7f46b. [DOI] [PubMed] [Google Scholar]
- 34.Drobna Z, Del Razo LM, Garcia-Vargas GG, Sanchez-Pena LC, Barrera-Hernandez A, Styblo M, et al. Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico. Journal of exposure science & environmental epidemiology. 2013;23:151–5. doi: 10.1038/jes.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beebe-Dimmer JL, Iyer PT, Nriagu JO, Keele GR, Mehta S, Meliker JR, et al. Genetic variation in glutathione S-transferase omega-1, arsenic methyltransferase and methylene-tetrahydrofolate reductase, arsenic exposure and bladder cancer: a case-control study. Environmental health : a global access science source. 2012;11:43. doi: 10.1186/1476-069X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampayo-Reyes A, Hernandez A, El-Yamani N, Lopez-Campos C, Mayet-Machado E, Rincon-Castaneda CB, et al. Arsenic induces DNA damage in environmentally exposed Mexican children and adults. Influence of GSTO1 and AS3MT polymorphisms. Toxicological sciences : an official journal of the Society of Toxicology. 2010;117:63–71. doi: 10.1093/toxsci/kfq173. [DOI] [PubMed] [Google Scholar]
- 37.Gong ZL, Lu XF, Cullen WR, Le XC. Unstable trivalent arsenic metabolites, monomethylarsonous acid and dimethylarsinous acid. J Anal Atom Spectrom. 2001;16:1409–13. [Google Scholar]
- 38.Le XC, Ma M. Short-column liquid chromatography with hydride generation atomic fluorescence detection for the speciation of arsenic. Analytical chemistry. 1998;70:1926–33. doi: 10.1021/ac971247q. [DOI] [PubMed] [Google Scholar]
- 39.Devesa V, Maria Del Razo L, Adair B, Drobna Z, Waters SB, Hughes MF, et al. Comprehensive analysis of arsenic metabolites by pH-specific hydride generation atomic absorption spectrometry. J Anal Atom Spectrom. 2004;19:1460–7. [Google Scholar]
- 40.Hernandez-Zavala A, Drobna Z, Styblo M, Thomas DJ. Analysis of arsenical metabolites in biological samples. In: Maines Mahin D., editor. Current protocols in toxicology. Vol. 42. 2009. pp. 4 33 1–4 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nermell B, Lindberg AL, Rahman M, Berglund M, Persson LA, El Arifeen S, et al. Urinary arsenic concentration adjustment factors and malnutrition. Environmental research. 2008;106:212–8. doi: 10.1016/j.envres.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Del Razo LM, Garcia-Vargas GG, Valenzuela OL, Castellanos EH, Sanchez-Pena LC, Currier JM, et al. Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapan and Lagunera regions in Mexico. Environmental health : a global access science source. 2011;10:73. doi: 10.1186/1476-069X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 44.Gardner RM, Engstrom K, Bottai M, Hoque WA, Raqib R, Broberg K, et al. Pregnancy and the methyltransferase genotype independently influence the arsenic methylation phenotype. Pharmacogenetics and genomics. 2012;22:508–16. doi: 10.1097/FPC.0b013e3283535d6a. [DOI] [PubMed] [Google Scholar]
- 45.Hopenhayn C, Huang B, Christian J, Peralta C, Ferreccio C, Atallah R, et al. Profile of urinary arsenic metabolites during pregnancy. Environmental health perspectives. 2003;111:1888–91. doi: 10.1289/ehp.6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilsner JR, Hall MN, Liu X, Ilievski V, Slavkovich V, Levy D, et al. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PloS one. 2012;7:e37147. doi: 10.1371/journal.pone.0037147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou WC, Chung YT, Chen HY, Wang CJ, Ying TH, Chuang CY, et al. Maternal arsenic exposure and DNA damage biomarkers, and the associations with birth outcomes in a general population from taiwan. PloS one. 2014;9:e86398. doi: 10.1371/journal.pone.0086398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moskvina V, Schmidt KM. On multiple-testing correction in genome-wide association studies. Genetic epidemiology. 2008;32:567–73. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- 49.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. American journal of human genetics. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamadani JD, Grantham-McGregor SM, Tofail F, Nermell B, Fangstrom B, Huda SN, et al. Pre- and postnatal arsenic exposure and child development at 18 months of age: a cohort study in rural Bangladesh. International journal of epidemiology. 2010;39:1206–16. doi: 10.1093/ije/dyp369. [DOI] [PubMed] [Google Scholar]
- 51.Schlebusch CM, Gattepaille LM, Engstrom K, Vahter M, Jakobsson M, Broberg K. Human adaptation to arsenic-rich environments. Molecular biology and evolution. 2015;32:1544–55. doi: 10.1093/molbev/msv046. [DOI] [PubMed] [Google Scholar]
- 52.Steinmaus C, Ferreccio C, Acevedo J, Yuan Y, Liaw J, Duran V, et al. Increased lung and bladder cancer incidence in adults after in utero and early-life arsenic exposure. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2014;23:1529–38. doi: 10.1158/1055-9965.EPI-14-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan Y, Marshall G, Ferreccio C, Steinmaus C, Selvin S, Liaw J, et al. Acute myocardial infarction mortality in comparison with lung and bladder cancer mortality in arsenic-exposed region II of Chile from 1950 to 2000. American journal of epidemiology. 2007;166:1381–91. doi: 10.1093/aje/kwm238. [DOI] [PubMed] [Google Scholar]
- 54.Smith AH, Marshall G, Yuan Y, Ferreccio C, Liaw J, von Ehrenstein O, et al. Increased mortality from lung cancer and bronchiectasis in young adults after exposure to arsenic in utero and in early childhood. Environmental health perspectives. 2006;114:1293–6. doi: 10.1289/ehp.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Navas-Acien A, Francesconi KA, Silbergeld EK, Guallar E. Seafood intake and urine concentrations of total arsenic, dimethylarsinate and arsenobetaine in the US population. Environmental research. 2011;111:110–8. doi: 10.1016/j.envres.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.