Figure 3.
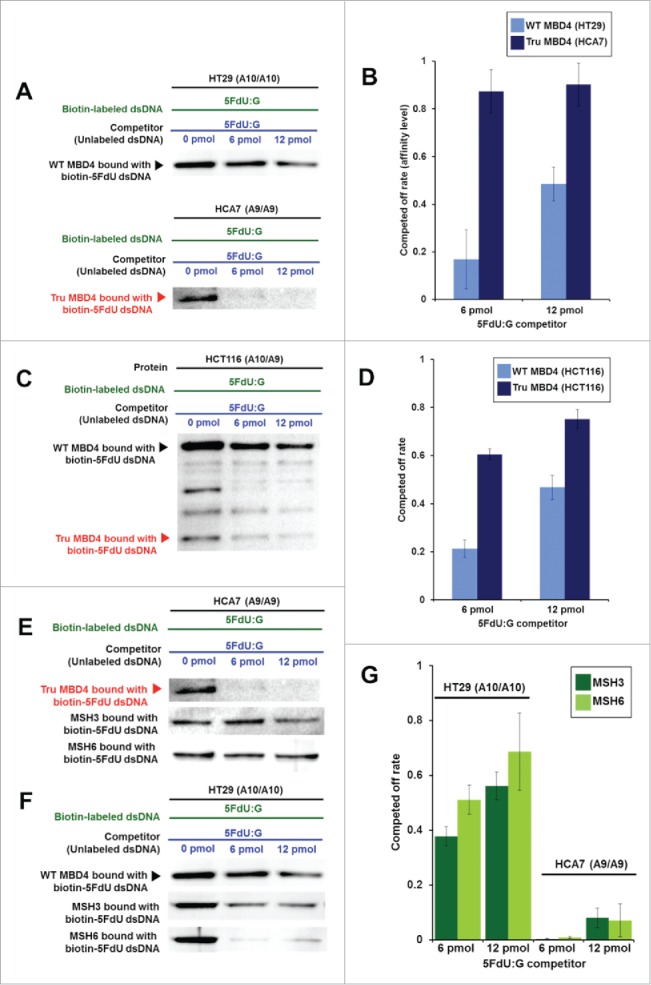
TruMBD4 binds to 5FU incorporated into DNA with higher affinity than normal MBD4 protein, and reduces 5FU affinity of DNA mismatch repair proteins in DNA pull down assays. (A) (Upper panel) Nuclear lysates from HT29 cells containing protein derived from wild type MBD4. As amounts of 5FdU:G competitor is increased relative to bound biotin-labeled 5FdU:G, the amount of MBD4 protein precipitated by biotin-labeled 5FdU:G is partially reduced, indicating some MBD4 protein was bound (or stolen) by the 5FdU:G competitor. This was not the case with a control complementary competitor, indicating MBD4 specifically recognizes 5FU within DNA (not shown). (Lower panel) Nuclear lysates from HT29 containing protein derived from frameshifted mutant MBD4 (TruMBD4). Here, as amounts of 5FdU:G competitor increased relative to biotin-labeled 5FdU:G, the amount of TruMBD4 precipitated by biotin-labeled 5FdU:G DNA is markedly reduced, suggesting a relative higher affinity for 5FdU:G as compared to normal MBD4 protein. (B) Bar graphs representing the reduction (competed off) rate by the protein for the 5FdU:G competitor, equating to the relative affinity level of the protein for 5FU within DNA. The affinity level of TruMBD4 for 5FdU:G is markedly higher than that of normal MBD4 protein. (C) Nuclear lysates from HCT116 cells containing both normal MBD4 protein and TruMBD4. As amounts of 5FdU:G competitor is increased, both normal MBD4 protein and TruMBD4 are competed off, but at apparently different rates. (D) Bar graph representing the competed off rate for both normal MBD4 protein and TruMBD4 from HCT116 cells. The relative affinity level of TruMBD4 was higher than normal MBD4 protein for the 5FdU:G competitor. (E, F) Nuclear lysates from HCA7 cells (E) and HT29 cells (F), demonstrating the relative pull down and competition off binding by MBD4/TruMBD4, MSH3 (key component of the hMutSβ MMR recognition complex) and MSH6 (key component of the hMutSα MMR recognition complex) for 5FdU:G. Note the relative difficulty for “compete off” bound reduction for MSH3 and MSH6 by the 5FdU:G competitor when TruMBD4 is present, compared to the “compete off” reduction for MSH3 and MSH6 when normal MBD4 protein is present. (F) Bar graph representing the competed off rate for MSH3 and MSH6 for the 5FdU:G competitor in the presence of normal MBD4 protein or TruMBD4. With normal MBD4 protein, MSH6 shows higher affinity for 5FU within DNA (as expected). However the binding affinity rates of both MMR proteins are markedly lower in TruMBD4-expressed cells than that of normal MBD4-expressed cells.
