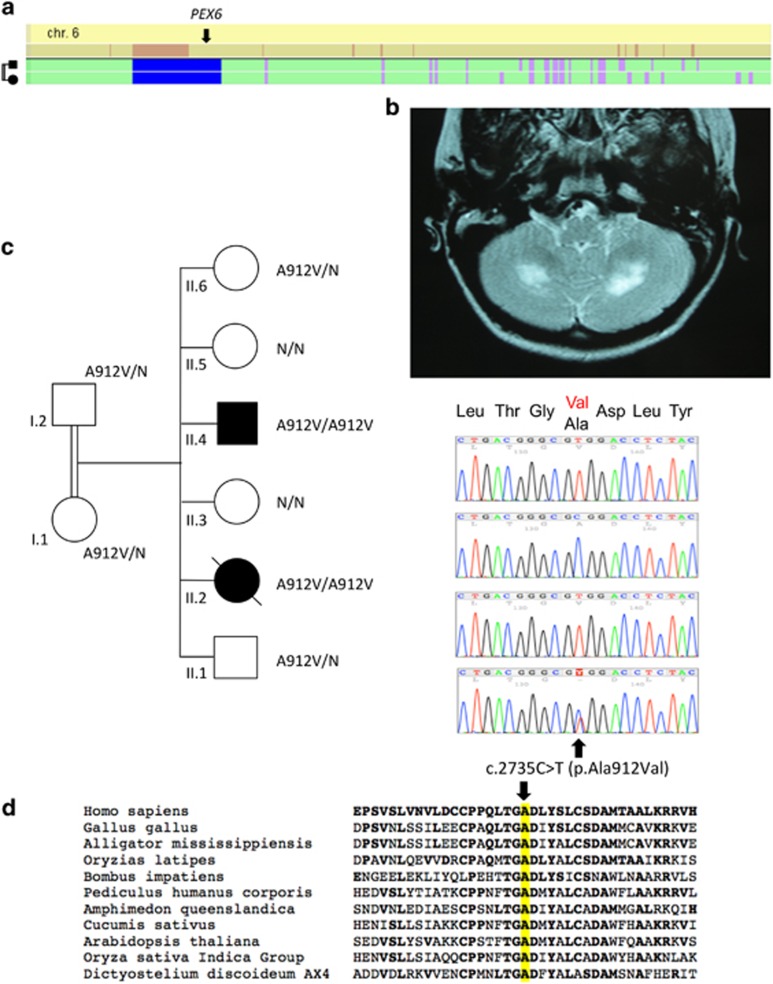
Figure 3.
Identification of PEX6 mutation in family C. (a) Whole-genome homozygosity mapping results. SNP genotyping results of the two patients were analyzed with the HomoSNP software in order to map homozygous regions across the 22 autosomes. Results are shown for chromosome 6, which was one of the seven chromosomes that revealed significant regions of homozygosity shared among the two affected children. Each colored line represents one patient, the pedigree is indicated on the left of the chromosome. Homozygous regions are colored according to the number of consecutive homozygous SNP markers: pink, from 20 to 24 markers; purple, from 25 to 29 markers; blue, 30 and more markers. Known loci for genes involved in ataxia are indicated in the line above the mapping results. The localization of PEX6 on the homozygosity map of chromosome 6 is indicated by an arrow. (b) T2-weighted horizontal MRI of patient II.2 at 13 years of age, showing hyper-echogenicity of the cerebellar white matter, suggesting demyelination of the afferent fibers. (c) Pedigree showing consanguinity and segregation of the disease with mutation c.2735C>T causing the p.(Ala912Val) missense change (A912V). Sanger sequencing results are indicated on the right of each corresponding sibling: both patients are homozygous for the mutation c.2735C>T and the parents are heterozygous for the same substitution. (d) Amino-acid sequence comparison of orthologous PEX6 proteins from different species. Amino acids that are identical to the human PEX6 sequence are shown in bold. The position of missense mutation (on the top of the mutated amino acid alanine, A, highlighted in yellow) is indicated by an arrow. The mutated amino acid is conserved in all eukaryotes including plants.