Abstract
Morphological integration predicts that correlated characters will coevolve; thus, each distinct suite of correlated characters might be expected to evolve according to a separate clock or ‘pacemaker’. Characters in a large morphological dataset for mammals were found to be evolving according to seven separate clocks, each distinct from the molecular clock. Total-evidence tip-dating using these multiple clocks inflated divergence time estimates, but potentially improved topological inference. In particular, single-clock analyses placed several meridiungulates and condylarths in a heterodox position as stem placentals, but multi-clock analyses retrieved a more plausible and orthodox position within crown placentals. Several shortcomings (including uneven character sampling) currently impact upon the accuracy of total-evidence dating, but this study suggests that when sufficiently large and appropriately constructed phenotypic datasets become more commonplace, multi-clock approaches are feasible and can affect both divergence dates and phylogenetic relationships.
Keywords: Mammalia, total-evidence dating, relaxed clocks, Bayesian phylogenetics, morphological integration, tip-dating
1. Introduction
Rates of molecular evolution typically vary across lineages, leading to the implementation of relaxed clock models in phylogenetics [1]. However, the advent of genomic scale data has revealed that different molecular partitions can exhibit different patterns of rate variation among lineages (heterotachy), requiring separate relaxed clocks. For example, owing to shared features of mutation, drift and selection, several mitochondrial genes might show a common pattern of heterotachy (e.g. all faster on the same branches) which is best modelled using a single relaxed clock separate to the relaxed clocks used for other (e.g. nuclear) genes, which might follow a different clock or ‘pacemaker’ [2].
ClockstaR [3] implements a computationally efficient approach for ascertaining the number of pacemakers in large datasets. It begins with a large number of candidate partitions, compares relative branch lengths for these partitions and then groups candidate partitions that share similar relative branch lengths, thus retrieving the minimum number of relaxed clocks that fit the data. While it has thus far only been used on genetic datasets, this approach is also highly relevant for large phenotypic/morphological datasets. Because of morphological integration [4,5], changes in certain traits will be accompanied by changes in others that are linked functionally, developmentally or genetically. These correlated traits should exhibit concordant patterns of evolution and rate variation [5], and thus share a common clock.
A ‘phenomic’ dataset for mammals [6] represents a promising morphological analogue for the genomic datasets recently subjected to multi-clock analyses [2]. It contains a sufficiently large number of traits (more than 4500) to test for the existence of subsets of characters each evolving under separate pacemakers. The ClockstaR analysis suggests that morphological trait evolution is governed by multiple relaxed clocks, none of which match the relaxed clock governing molecular evolution. In total-evidence tip-dating [7], which uses the ages and morphological characters of fossil taxa to infer divergence dates and evolutionary rates [1,8], using these multiple relaxed clocks—rather than the typical single global clock [7], which is likely a poor fit [8]—results in major differences to inferred divergence dates and tree topology, although these differences are not always in the direction anticipated (i.e. a seemingly more appropriate clock model yields less plausible dates).
As detailed in Results and Discussion, there are caveats that mean the empirical results of these analyses should be treated with caution. Rather, this study should be seen as a proof-of-concept demonstration that multi-clock analyses can be implemented on sufficiently large phenotypic datasets, first to circumscribe modules of correlated characters, and then to use an appropriate number of relaxed clocks in phylogenetic analysis, which can significantly influence topology and divergence dates.
2. Methods
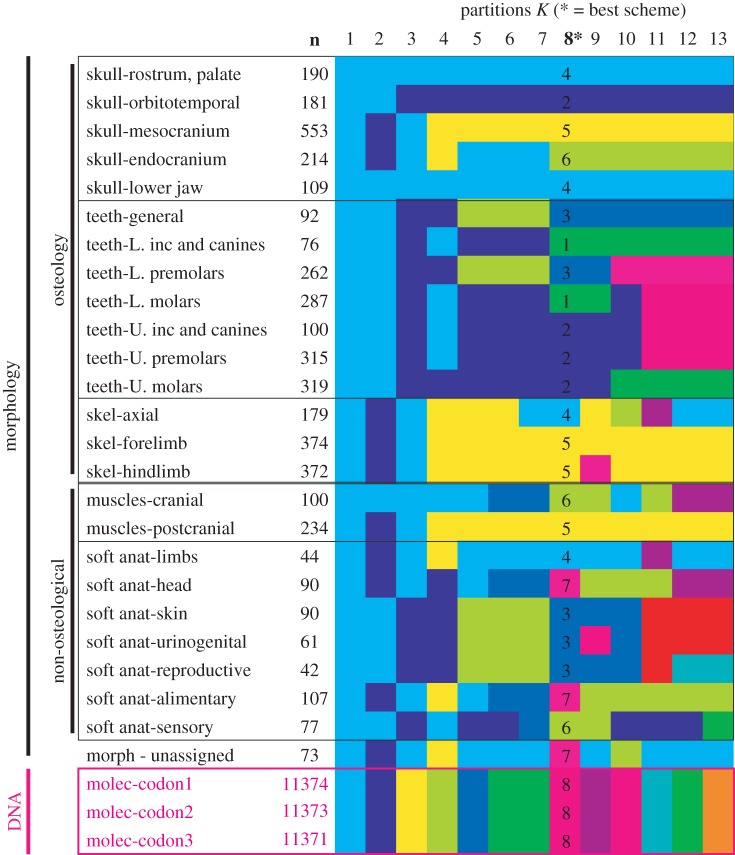
The data matrix [6] consisted of 86 mammal species scored for 4541 morphological characters and 27 genes (36 860 nucleotides). The 4541 morphological characters were divided into 25 candidate partitions (Dryad file A1; see the electronic supplementary material) based on anatomical area and type of character (e.g. forelimb skeleton and cranial myology), with each candidate partition having sufficient characters (42–553) to potentially enable good estimates of branch lengths. ClockstaR (v. 2.0.1) analyses (files B1–B7) ideally require branch lengths for all candidate character partitions and thus, all taxa should ideally have some data for every character partition. For this reason, only the 46 extant taxa were considered (the fossils lacking all candidate genetic partitions, and often also many candidate morphological partitions). Furthermore, the distribution of missing data (different genes missing for different taxa) meant that dividing the genetic data into candidate partitions by gene would have meant most partitions are completely missing data for several taxa; for this reason (and because the primary aim was not to investigate intergene variability), the genetic data were divided by codons into three candidate partitions (each with data for every taxon; introns were excluded). ClockstaR requires an input tree topology; two rather different trees—constructed using different data and methods—were employed [6,9]. Branch lengths for all partitions on this input topology were ascertained using MrBayes [10], and the optimal number of partitions were identified by finding the global maximum in ClockstaR [3] using the FANNY algorithm, which is the only clustering method available which is ‘fuzzy’ (i.e. takes into consideration uncertainty in group membership). The ‘hard’ algorithms PAM and CLARA consistently favoured the maximum or minimum number of partitions, and were thus not employed; the reasons for this behaviour deserve further investigation. Both input trees returned an optimal scheme of approximately eight clock-partitions of similar composition; subsequent analyses used the results from the first tree. Randomized partitions of morphological characters yielded different schemes, favouring very many or very few partitions (files B8–10).
Dated total-evidence analyses of all 86 fossil and living taxa were then performed in MrBayes [10]. The Mk model was used for the morphological data, sampled-ancestor birth–death tree prior was employed, and two nodes (root, stem-therians: figure 2) were calibrated and enforced as monophyletic [11], all other ages and relationships were free to vary. PartitionFinder [12] was used to identify the substitution models for different subsets of the molecular data, using every codon for every locus as a candidate partition and the BIC with unlinked branch lengths, which is most conservative in estimating the number of required partitions. Two analyses were performed, using: a single clock (IGR) for the entire morphological and molecular dataset, or the multiple clocks identified by ClockstaR. The executable files for both analyses, with all prior, model and MCMC run settings are in files C1–11, along with convergence exploration in Tracer, AWTY and MrBayes.
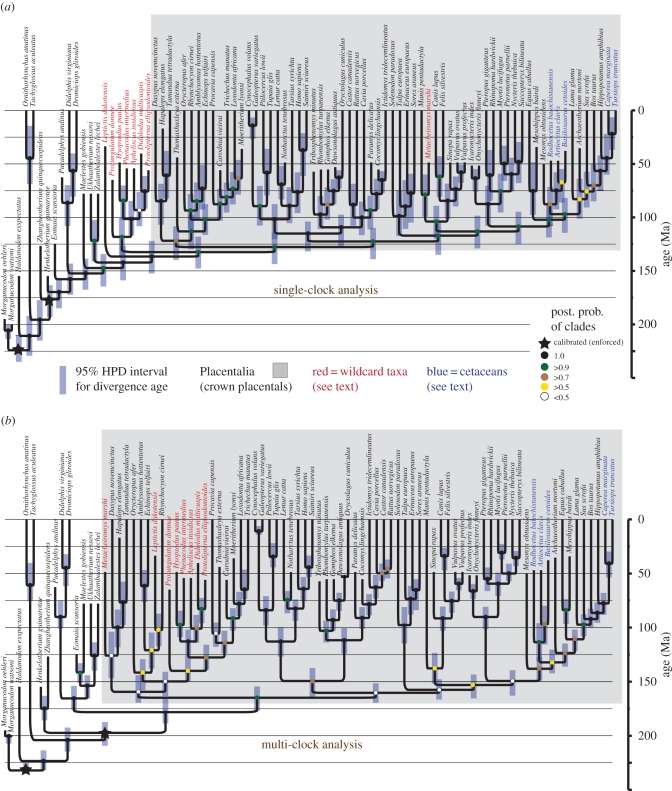
Figure 2.
Bayesian total-evidence tip-dated trees of mammals using (a) a single clock for all morphological and molecular traits, and (b) eight separate clocks (seven for morphology; one for DNA) with groupings as per figure 1. Note in (a) the anomalously basal position of many taxa (red) traditionally considered crown placentals, but also the shallower divergence dates (tabled in file C12). Posterior probabilities are colour-coded; exact values are presented in files C4 and C9. (Online version in colour.)
3. Results and discussion
The ClockstaR analyses suggested the existence of eight clocks, with the morphological traits evolving according to seven separate pacemakers, whereas the candidate molecular partitions evolved under one common pacemaker (figure 1). Notably, no morphological partition was considered to share the (single) molecular clock [8]: if this molecular clock is assumed to be relatively rate-constant (as suggested by molecular branch lengths: file B3), then it implies that all morphological partitions deviate from strict-clock-like behaviour. However, they appear to deviate in predictable clusters: many of the groups of characters identified as sharing one of the separate clocks appear biologically reasonable. In the optimal eight-clock scheme, and schemes with slightly fewer or more clocks, single clocks characterize (i) all the dental traits in the upper jaw, (ii) the skeletal traits of the forelimb and hindlimb plus postcranial muscles and (iii) the rostrum, palate and the lower jaw. However, other groupings are not particularly intuitive: for instance, the mesocranium and endocranium are split across separate clocks, with the mesocranium sharing the aforementioned ‘limb’ clock. Similarly, the cranial soft anatomy and alimentary systems consistently share the same clock. Further investigation is required to test whether these unexpected shared clocks are due to stochastic factors (relatively few characters per partition, compared with genomic data), genuine biological correlation (e.g. developmental timing rather than anatomical proximity), or are artefacts of character selection (see below).
Figure 1.
The 28 candidate clock partitions (25 morphological, three molecular) and their optimal grouping patterns when assigned multiple clocks from n = 1, 2, 3, 4, …, 13 clocks or pacemakers (optimal number = 8). Note that the three molecular partitions consistently form a single clock, separate from all morphological partitions. Some groups of morphological partitions also consistently fall within the same clock (e.g. upper teeth; forelimb and hindlimb skeleton; skin, urinogenital and reproductive systems; soft anatomy of head and alimentary systems). (Online version in colour.)
For the phylogenetic analyses, PartitionFinder identified seven separate nucleotide models and partitions; these showed consistent groupings by codon and/or gene, e.g. some partitions consisted entirely of third codons for different genes, others consisted of different codons for a single gene (file C1). The dated total-evidence analyses with MrBayes using these partitions and models yielded convergence diagnostics which were strong for the single-clock analysis (files C5–6). For the multi-clock analysis, convergence was less certain (files C10–11); however, each separate MCMC run gave near identical trees and divergences. The good convergence for the single-clock analysis was notable, given that less parametrized (i.e. undated) total-evidence Bayesian analyses previously did not converge [6]. This might be due to the smaller number of molecular substitution models and partitions employed here, and additional topology moves for mixed datasets introduced in newer versions of MrBayes. Thus, despite high computational demands, total-evidence tip-dating can be applied successfully to relatively large datasets encompassing thousands of phenotypic characters and dozens of genes.
The phylogenies from the single-clock and multi-clock Bayesian analyses (figure 2) exhibit major differences in dates as well as topology, demonstrating that multi-clock models impact both areas. Surprisingly, the single-clock model generates divergence dates that are more congruent with other analyses, e.g. the common ancestor of placentals is dated at approximately 123 Ma rather than approximately 170 Ma. However, both these total-evidence dates for placental divergences are likely to be overestimates to some extent [9,11]; analyses of huge genomic datasets with robust node calibrations have retrieved ages of approximately 90 Ma [9]. Thus, use of an (arguably) more appropriate multi-clock model [8] exaggerates rather than ameliorates the anomalously deep divergence dates generated by total-evidence analyses. Possible reasons are discussed below.
Conversely, the topology of the multi-clock tree is generally more consistent with recent studies and broadly accepted views [6,13]. In particular, the positions of several fossils are resolved more plausibly (figure 2). The South American ‘meridiungulates’ (liptoternans Didolodus, Protolipterna) are crown placentals: they fall most frequently near afrotherians, but also sometimes near ungulates, resulting in low clade support across the base of mammals (figure 2b). These alternative positions within crown placentals mirror morphological studies [6,14] as well as independent evidence from ancient proteins [15]. Similarly, three condylarth-grade taxa (Hyopsodus, Apheliscus and Phenacodus) are retrieved within crown placentals near afrotherians or ungulates, in agreement with most other studies [6,13]. In contrast, the single-clock tree places these meridiungulates and condylarths in a heterodox position outside of crown placentals altogether.
Other differences between the multi- versus single-clock trees mirror ongoing debates [6,13]: Protungulatum is either a crown or a stem placental, Leptictis is either an atlantogenatan relative or a stem placental, and Metacheiromys is either related to edentates or to pangolins. It seems that for each of these questions, morphology contains two strong alternative signals, and the choice of clock model determines which predominates. Finally, the (likely artefactual) diphyly of cetaceans generated by this dataset in undated analyses [6] is replicated in both clock analyses, with primitive fossil whales (scored only for morphology) falling near mesonychids as basal cetartiodactylans, and living cetaceans (with characters dominated by DNA) grouping with hippos.
There are important caveats that should be emphasized. Most notably, character choice (over/undersampling of characters along certain branches) will impact upon fit of multi-clock models, as well as the resultant total-evidence phylogenetic analyses. For instance, the substitution trees for the candidate morphological partitions consistently differ from the candidate molecular partitions in having shorter terminal branch lengths (file B6), which is likely to be at least partly owing to sampling (ascertainment) bias. While the dataset analysed differs from most morphological datasets in explicitly sampling autapomorphies [6], it might not have sampled them as intensely as changes on internal branches, leading to artefactually truncated terminal branches for the morphological partitions. Similar uneven sampling across branches for different subsets of morphological data might also generate spurious clusters and divisions of characters when multi-clock analyses are implemented. Such uneven sampling will also impact upon total-evidence dating, which is most accurate when changes across all branches are sampled with similar intensity (as in molecular data). A fully objective quantification of the amount of morphological change on any (and all) branches will be challenging, given the inevitable subjectivity of atomizing phenotypic differences into characters and character states. However, it should be easy to vastly improve upon the current widespread practice of totally ignoring changes on terminal branches when constructing cladistic data matrices: the relatively simple procedure of sampling autapomorphies with the same zeal as other characters would greatly improve the suitability of morphological datasets for clock analysis. Until morphological datasets gathered in this way can begin to be analysed, studies inferring numbers of morphological clocks, and total-evidence dating, should be treated with caution. Biased selection of characters, as well as episodic evolution of morphological traits [8,11], could be at least partly driving overestimates of divergence times in tip-dating analyses.
These issues notwithstanding, the current results demonstrate that—with large character sets [6,16] that are sampled appropriately (see above)—it is possible to use molecular clock methods to identify suites of morphological traits that are evolving according to common pacemakers [3]. These patterns, in combination with more mechanistic approaches, will together help elucidate the genetic, developmental, functional and ecological processes generating integrated evolutionary modules [4,5]. Furthermore, such improvements on our currently inadequate models of morphological evolution could ameliorate some of the issues potentially affecting the reliability of tip- and total-evidence dating [8,11]. Using the appropriate number of clocks in such analyses, rather than a single clock, can substantially influence retrieved topologies and divergence dates.
Supplementary Material
Acknowledgements
I thank eResearch SA (eRSA) for high-performance computing facilities; and T. Guillerme, A. Wright and an anonymous reviewer for constructive comments.
Data accessibility
All data files are described in the electronic supplementary material and are available from Dryad at: http://dx.doi.org/10.5061/dryad.3h4m5.
Authors' contributions
M.L. conceived the study, performed the analyses, interpreted the results, wrote the paper and agrees to be accountable for this work.
Competing interests
I have no competing interests.
Funding
Australian Research Council grant no. DP160103005.
References
- 1.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88 ( 10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duchêne S, Ho SYW. 2015. Mammalian genome evolution is governed by multiple pacemakers. Bioinformatics 31, 2061–2065. ( 10.1093/bioinformatics/btv121) [DOI] [PubMed] [Google Scholar]
- 3.Duchêne S, Molak M, Ho SYW. 2014. ClockstaR: choosing the number of relaxed-clock models in molecular phylogenetic analysis. Bioinformatics 30, 1017–1019. ( 10.1093/bioinformatics/btt665) [DOI] [PubMed] [Google Scholar]
- 4.Olson EC, Miller RL. 1958. Morphological integration. Chicago, IL: University of Chicago Press. [Google Scholar]
- 5.Clarke JA, Middleton KM. 2008. Mosaicism, modules, and the evolution of birds: results from a Bayesian approach to the study of morphological evolution using discrete character data. Syst. Biol. 57, 185–201. ( 10.1080/10635150802022231) [DOI] [PubMed] [Google Scholar]
- 6.O'Leary MA, et al. 2013. The placental mammal ancestor and the post-K–Pg radiation of placentals. Science 339, 662–667. ( 10.1126/science.1229237) [DOI] [PubMed] [Google Scholar]
- 7.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Reilly J, dos Reis M, Donoghue PCJ. 2015. Dating tips for divergence time estimation. Trends Genet. 31, 637–650. ( 10.1016/j.tig.2015.08.001) [DOI] [PubMed] [Google Scholar]
- 9.dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. B 279, 3491–3500. ( 10.1098/rspb.2012.0683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck RMD, Lee MSY. 2014. Ancient dates or accelerated rates? Morphological clocks and the antiquity of placental mammals. Proc. R. Soc. B 281, e20141278 ( 10.1098/rspb.2014.1278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 13.Halliday TJD, Upchurch P, Goswami A. 2016. Resolving the relationships of Paleocene placental mammals. Biol. Rev. ( 10.1111/brv.12242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agnolin FL, Chimento NR. 2011. Afrotherian affinities for endemic South American ‘ungulates’;. Mammal Biol. 76, 101–108. ( 10.1016/j.mambio.2010.12.001) [DOI] [Google Scholar]
- 15.Welker F, et al. 2015. Ancient proteins resolve the evolutionary history of Darwin's South American ungulates. Nature 522, 81–84. ( 10.1038/nature14249) [DOI] [PubMed] [Google Scholar]
- 16.Ni X, Gebo DL, Dagosto M, Meng J, Tafforeau P, Flynn JJ, Beard KC. 2013. The oldest known primate skeleton and early haplorhine evolution. Nature 498, 60–64. ( 10.1038/nature12200) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data files are described in the electronic supplementary material and are available from Dryad at: http://dx.doi.org/10.5061/dryad.3h4m5.