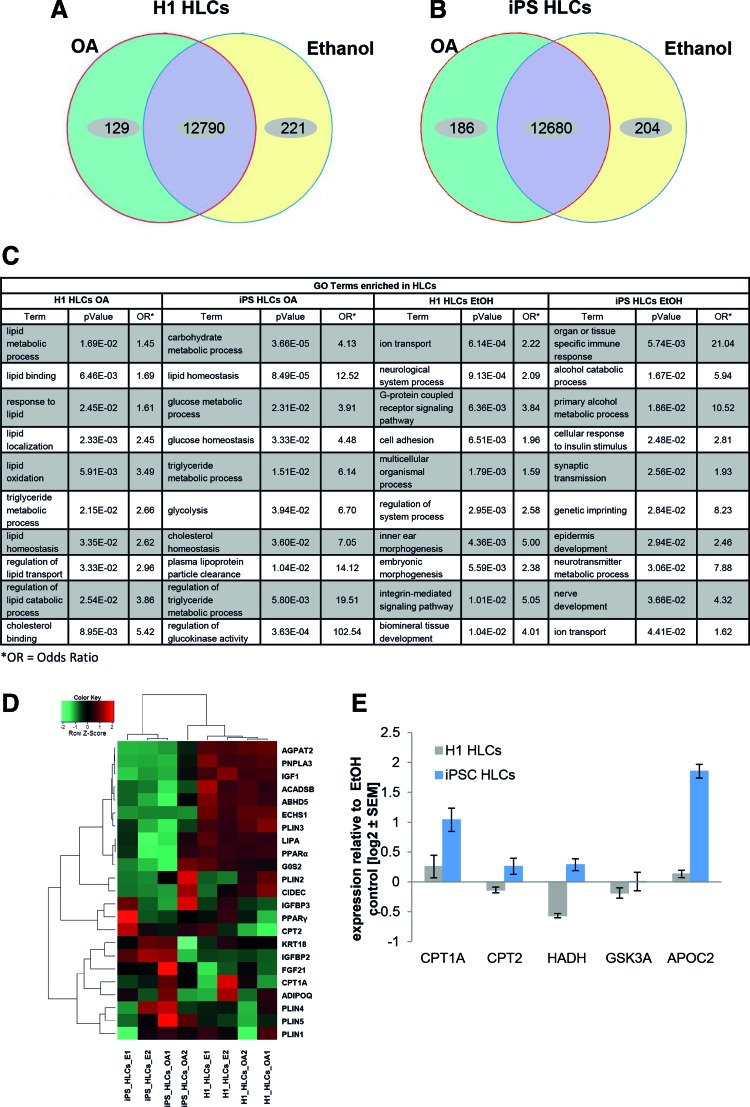
FIG. 4.
Gene expression profiles of HLCs challenged with OA enable characterization of the early steps in NAFLD. Venn diagrams of genes expressed (detection P value <0.05) in H1- (A) or iPSC- (B) derived HLCs (n = 2). Green and yellow segments represent genes exclusively expressed in OA treated or control cells, respectively. Purple segments comprise genes, which are expressed under both conditions, but not necessarily with the same intensity. (C) GO-analysis of genes upregulated or exclusively expressed under OA treatment reveals an enrichment of NAFLD-related categories, while ethanol control cells predominantly express genes mapping to divergent categories. Shown are preselected significant GO-Terms; for full data set, see Supplementary Tables S6 and S7. (D) Heatmap of genes involved in insulin signaling and lipid or glucose metabolism reveals differences between H1- and iPSC-derived HLCs, which might, in part, be due to their distinct genetic background. However, qualitative changes are similar between both groups, which are reflected by the formation of subclusters connected to OA treatment. (E) Expression of genes involved in lipid and glucose metabolism was analyzed using qRT-PCR (n = 2). Gene expression was normalized to β-actin and, subsequently, to the control samples. For each sample, the mean ± standard error of duplicate experiments is shown as log2 scale. GO, gene ontology; HLCs, hepatocyte-like cells; iPSC, induced pluripotent stem cell; NAFLD, nonalcoholic fatty liver disease; OA, oleic acid; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd