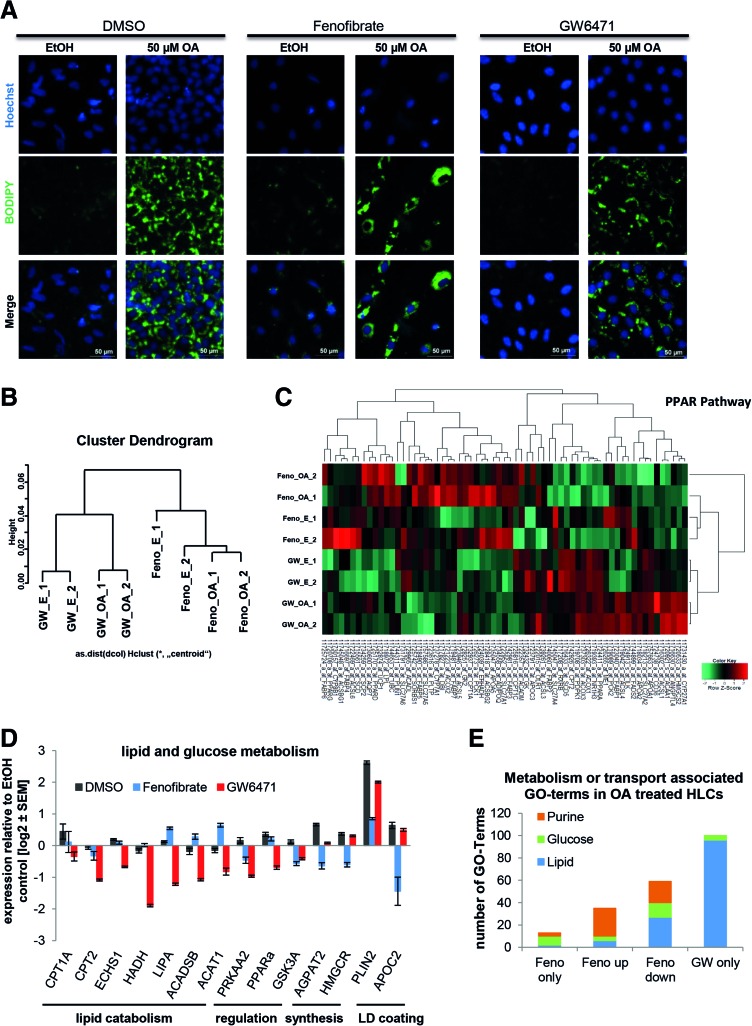
FIG. 7.
PPARα signaling has major impact on the induction of steatosis. IPSC-derived HLCs were incubated in parallel with either Fenofibrate (Feno, PPARα agonist) or GW6471 (PPARα antagonist) during the 48 h OA induction. (A) Steatosis induction was monitored by BODIPY 493/503 (green) staining of LDs. In every case, LDs increased after OA induction (right columns) compared to the control (left columns), but no differences between the different PPARα treatments are visible. (B) Microarray-based transcriptome analysis revealed two distinct clusters representing Fenofibrate and GW6471 treatment and two subclusters related to OA treatment (n = 2). (C) Heatmap representation of PPAR pathway genes shows distinct gene expression profiles for Fenofibrate (“F”) and GW6471 (“GW”) treated cells with qualitative changes after OA induction. Expression of genes involved in lipid or glucose metabolism (D) was analyzed using qRT-PCR (n = 2). As expected, in most cases, Fenofibrate and GW6471 treatment had opposing effects on gene expression. Gene expression was normalized to β-actin and, subsequently, to the ethanol-treated control samples. For each sample, the mean ± standard error of duplicate experiments is shown as log2 scale. (E) Significantly expressed genes from the global analysis were subdivided into genes only expressed after Fenofibrate treatment, up- or downregulated after Fenofibrate treatment, compared to GW6471 treatment or only expressed after GW6471 treatment. Then they were assigned to GOs (Supplementary Table S9). The numbers of GO-terms that were associated with lipid, glucose, or purine metabolism and transport are displayed. GO, gene ontology; HLCs, hepatocyte-like cells; LDs, lipid droplets; OA, oleic acid; PPARα, peroxisome proliferator-activated receptor alpha; qRT-PCR, quantitative reverse transcription polymerase chain reaction. Color images available online at www.liebertpub.com/scd