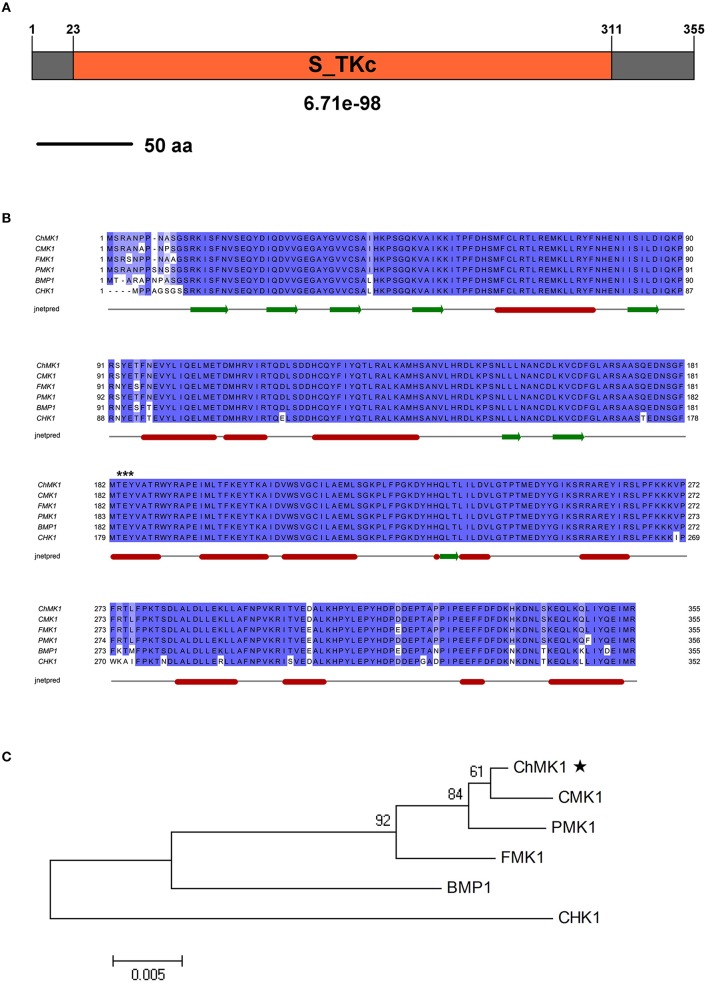
Figure 1.
Characterization of the C. higginsianum ChMK1 protein. (A) The domain structure of C. higginsianum ChMK1 as annotated by SMART MODE (http://smart.embl-heidelberg.de/smart/change_mode.pl). (B) Alignment analysis of the amino acid sequences of ChMK1 (CH063_08490) and its homologs from other fungi using Clustal X program. PMK1: XP_003712175.1 (M. oryzae); CHK1: AF178977.1 (C. heterostrophus); BMP1: AF205375.1 (B. cinerea); FMK1: AAG01162.1 (F. oxysporum); CMK1: AAD50496.1 (C. lagenarium). The conserved kinase activation residues TEY are labeled with asterisks at the top of the alignment. The secondary structure prediction was completed with Jnetpred program. The green arrows indicate β-strands structure and the red sticks indicate α-helix structure. (C) Phylogenetic analysis of ChMK1 of C. higginsianum and its homologs from other fungi. The amino acid sequences were analyzed by MEGA version 4 with Unrooted Neighbor-joining algorithm. Bootstrap values were calculated from 1000 bootstrap replicates. Only bootstrap support values >50% are shown. The black star indicates ChMK1.