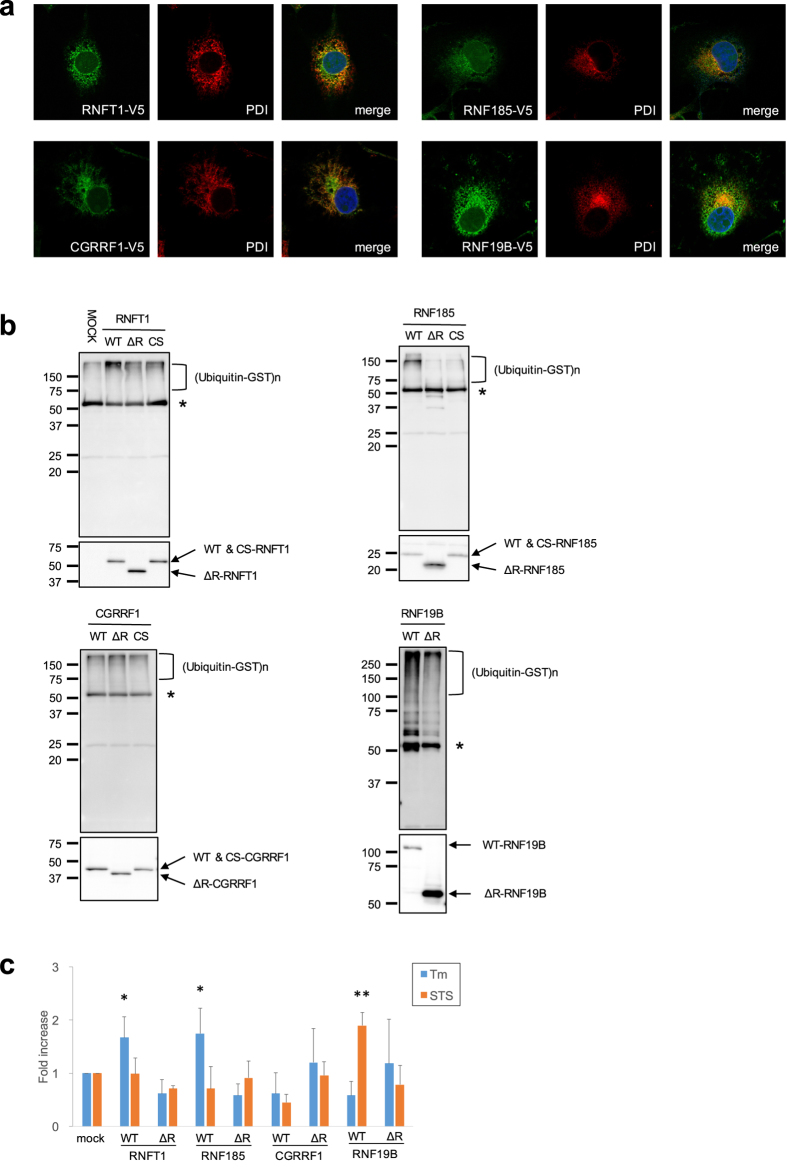
Figure 3.
Characterization of candidates for ERAD E3 ligase (a) Subcellular localisation of E3 ligases. COS-1 cells stably expressing E3-V5 were subjected to immunofluorescence staining with anti-V5 and -PDI antibodies. (b) In vitro autoubiquitination assay. The E3 proteins produced by a transcription/translation system were immunoprecipitated with anti-V5 antibody, and then mixed in the reaction buffer with E1 (GST-tagged), E2 (GST-UbcH5c) and HA-ubiquitin. The reaction mixture was again immunoprecipitated with anti-V5 antibody and analysed via Western blotting using anti-ubiquitin or V5-antibodies. CS mutants defective in E3 activity were constructed by replacement of conserved coordinating Cys with Ser residues in the RING. Asterisk indicates the heavy chain of immunoglobulin. (c) E3 ligases protect against ER stress-induced cell death. Neuro-2a (N2a) cells stably expressing WT or ΔRING (ΔR) mutant of E3 ligases were transiently treated with Tm (1 μg/ml) or staurosporine (STS; 0.1 μM) and incubated for 48 h. The cells were stained with crystal violet. The eluted dye at an optical density of 590 nm was measured. Cell viability was calculated as follows: OD for assay/OD for vehicle control (0.1% dimethyl sulfoxide) well. The results are expressed as the fold increase compared with mock cells, in terms of means ± SD (three independent experiments in duplicate). Statistical analysis was performed using ANOVA followed by Bonferroni correction (vs. controls; *p < 0.05, **p < 0.01)