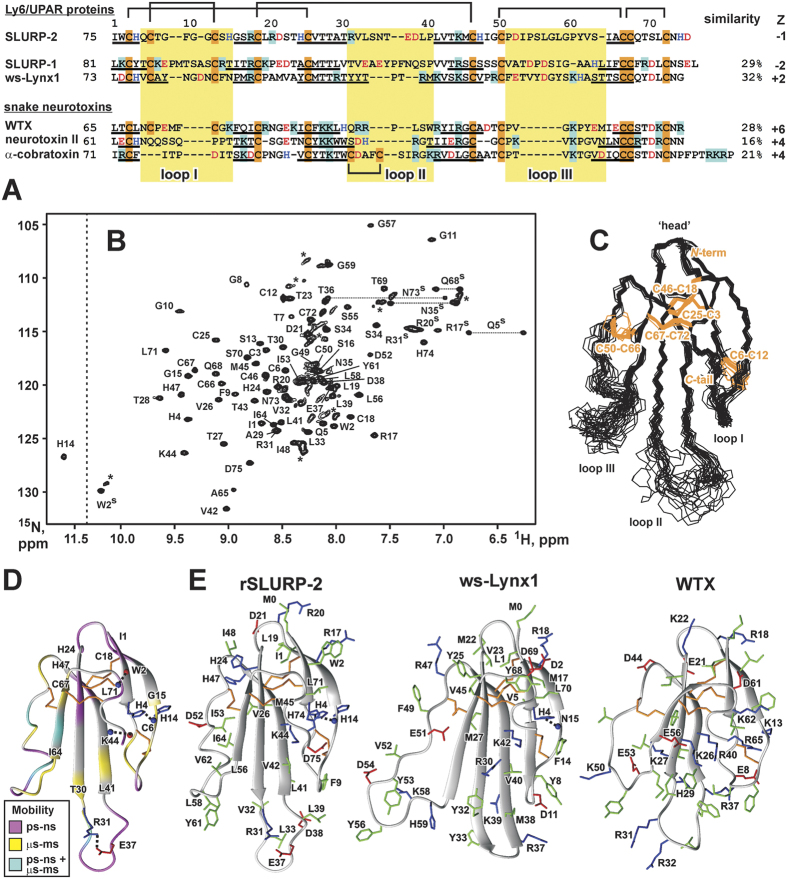
Figure 1. Amino acid sequence alignment and comparison of the spatial structure of SLURP-2 with other human Ly-6/uPAR proteins and three-finger snake neurotoxins.
(A) The sequence of the water-soluble domain of human Lynx-1 (ws-Lynx1) is shown without the GPI consensus sequence at the C-terminus. The positively charged (Arg/Lys), negatively charged (Asp/Glu), and His and Cys residues are highlighted. The fragments corresponding to β-strands in the spatial structures of the proteins are underlined. The loop regions are highlighted with a yellow background. (B) The 2D 1H,15N-HSQC spectrum of rSLURP-2 (0.5 mM, 5% dioxane, pH 5.0, 37 °C). The obtained resonance assignments are shown. The resonances of the side-chain groups are indicated by a superscripted “s”. The signals corresponding to the unfolded/aggregated protein are marked by asterisks. The relative population of this protein form did not exceed 10%. (C) The set of the best 20 rSLURP-2 structures were superimposed over the backbone atoms in regions with a well-defined structure. The three loops and ‘head’ of the protein are labeled. Disulfide bonds are shown in orange. (D) Ribbon representation of the spatial structure of rSLURP-2 with mapped dynamic NMR data. 15N relaxation rates were measured at 60 MHz (pH 5.0, 37 °C) for 0.08 mM rSLURP-2 in water without the addition of dioxane (Supplementary Fig. S4). The backbone fragments affected by dynamic processes on the ps-ns time scale (heteronuclear NOE < 0.7) or μs-ms time scale (R1 · R2 product > 20 s−2 or HN signals broadened beyond the detection limit) are highlighted. Additional electrostatic and hydrogen bonding interactions that stabilize the protein fold are shown. Backbone amide and carbonyl groups are indicated by blue and red spheres, respectively. (E) Comparison of the spatial structures of rSLURP-2, ws-Lynx1 and the WTX[P33A] mutant. Aromatic/hydrophobic, positively charged (including His), negatively charged, and Cys residues are indicated in green, blue, red, and orange, respectively.