The crystal structures of two salts of unsymmetrical dimethyl hydrazine show different protonation patterns of the cation.
Keywords: crystal structure, unsymmetrical dimethylhydrazine, protonation pattern, hydrogen bonds
Abstract
We describe the syntheses and crystal structures of two molecular salts containing the 1,1-dimethylhydrazinium cation, namely 1,1-dimethylhydrazin-1-ium bromide, C2H9N2 +·Br−, (I), and 2,2-dimethylhydrazin-1-ium dihydrogen phosphite, C2H9N2 +·H2PO3 −, (II). In (I), the cation is protonated at the methylated N atom and N—H⋯Br hydrogen bonds generate [010] chains in the crystal. In (II), the cation is protonated at the terminal N atom and cation-to-anion N—H⋯O and anion-to-anion O—H⋯O hydrogen bonds generate (001) sheets.
Chemical context
Unsymmetrical dimethylhydrazine (1,1-dimethylhydrazine; C2H8N2; UDMH) is a colourless liquid at room temperature and pressure with a strong and unpleasant ammonia-like or fishy smell. The best known application of this compound is the fuel (reducing agent) in hypergolic rocket fuels (Edwards, 2003 ▸), where it can be used alone or mixed with hydrazine: the latter formulation (trade name ‘Aerozine 50’) was used by the Apollo lunar modules to begin their homeward journeys from the moon.
Chemically, both nitrogen atoms in UDMH bear lone pairs of electrons, which can act as weak bases to accept protons and therefore result in the formation of molecular salts when reacted with acids. The first crystal structure of a UDMH salt was reported by Klapötke et al. (1999 ▸), who prepared 1,1-dimetylhydrazinium azide as a possible high-energy-density material with military applications; the methylated UDMH nitrogen atom is protonated and the components are linked by strong N—H⋯N hydrogen bonds in the crystal. However, this salt exhibited pronounced hygroscopic behaviour and had a low melting point of 311 K, which deemed it unsuitable for such uses. The nitrate salt of UDMH, which may be a decomposition product of hypergolic fuels, was prepared soon afterwards by the same workers (De Bonn et al., 2001 ▸) by a low-temperature, non-aqueous synthesis: anhydrous nitric acid and UDMH were separately dissolved in dichloromethane at 195 K and the solutions mixed at the same temperature. The resulting hygroscopic salt, 1,1-dimethylhydrazinium nitrate, is protonated at the methylated nitrogen atom and features N—H⋯O hydrogen bonds in its crystal structure.
Merkoulov et al. (2005 ▸) synthesized 1,1-dimethylhydrazinium chloride by reacting liquid UDMH with HCl dissolved in diethyl ether: its crystal structure consists of two independent cations and two chloride anions in the asymmetric unit. The cation is protonated at the methylated nitrogen atom and a dense network of strong N—H⋯Cl and weak C—H⋯Cl hydrogen bonds helps to consolidate the packing in the crystal. A salt with a more complicated counter-ion was synthesised by Mu et al. (2011 ▸): the addition of liquid UDMH to a solution of picric acid in ethanol at room temperature yielded 1,1-dimethylhydrazinium picrate. As before, the UDMH protonates at the methylated nitrogen atom and cation-to-anion N—H⋯O hydrogen bonds help to establish the packing.
As an extension of these studies, we now describe the syntheses and crystal structures of 1,1-dimethylhydrazin-1-ium bromide, C2H9N2 +·Br− (I) and 2,2-dimethylhydrazin-1-ium dihydrogen phosphite, C2H9N2 +·H2PO3 − (II).
Structural commentary
Compound (I) crystallizes in space group I2/a (non-standard setting of C2/c) with one cation and one bromide anion in the asymmetric unit (Fig. 1 ▸). The cation is protonated at the central N2 atom, as seen in previous UDMH salts referred to above. The N1—N2 bond length [1.4478 (19) Å] is slightly shorter than the C—N bond lengths [1.482 (2) and 1.485 (2) Å]. N2 is displaced from N1, C1 and C2 by 0.4834 (16) Å and the C—N—C bond angle [111.38 (14)°] is slightly greater than the C—N—N angles [108.93 (12) and 108.97 (14)°]. The H atoms attached to N1 point away from the carbon atoms [C1—N2—N1—H2n = −175.7 (2); C2—N2—N1—H1n = 178.0 (2)°] and the N2—H3n bond bisects the N1H2 group [H3n—N2—N1—H1n = 61 (2)°].
Figure 1.
The molecular structure of (I), showing 50% displacement ellipsoids. The N—H⋯Br hydrogen bond is indicated by a double-dashed line (Table 1 ▸).
Compound (II) crystallizes in space group Pna21 with one cation and one dihydrogen phosphite anion in the asymmetric unit (Fig. 2 ▸). In this case, the cation is protonated at the terminal N atom rather than the central N atom, which has not been seen previously in UDMH salts. The N1—N2 bond length is 1.454 (3) Å and the C—N bond lengths are 1.462 (3) and 1.463 (3) Å. The geometry about N2 is pyramidal and this atom is displaced from N1, C1 and C2 by 0.504 (2) Å. The bond angles about N2 show the same trend as those in (I): C—N—C = 110.69 (18); C—N—N = 107.62 (17) and 107.94 (18)°. Two of the H atoms attached to N1 have almost the same locations as the corresponding atoms in (I), whereas the third bisects the C1—N2—C2 grouping [C1—N2—N1—H3n = −62°]. In the anion, the P1—O3 bond length of 1.5638 (16) Å is typical (Harrison, 2003 ▸) for the protonated O atom in a dihydrogen phosphite group whereas P1—O1 [1.4982 (15) Å] and P1—O2 [1.5003 (16) Å] are almost the same length, indicating the expected delocalization (resonance) of the negative charge over these two O atoms. The O—P—O bond angle for the unprotonated oxygen atoms [116.76 (9)°] is significantly larger than the O—P—OH angles [106.37 (9) and 111.46 (9)°], as seen previously for similar species (Harrison, 2003 ▸). P1 is displaced from its attached O atoms by 0.4510 (13) Å.
Figure 2.
The molecular structure of (II), showing 50% displacement ellipsoids. The N—H⋯O hydrogen bond is indicated by a double-dashed line (Table 2 ▸).
Supramolecular features
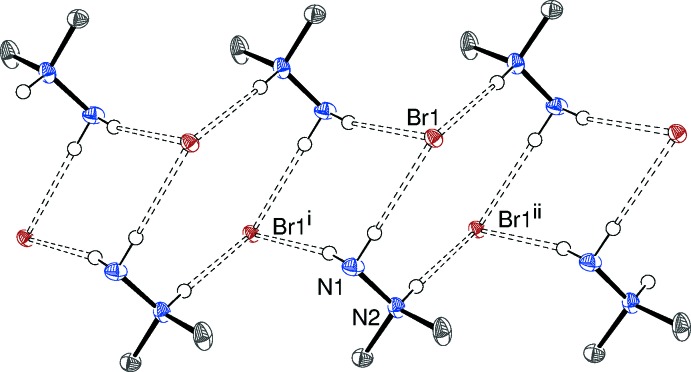
In the crystal of (I), N—H⋯Br hydrogen bonds (Table 1 ▸) link the components into [010] chains (Fig. 3 ▸): each Br− ion accepts three N—H⋯Br bonds and alternating, centrosymmetric  (8) and
(8) and  (10) loops occur within the chain. The N2 bond is significantly shorter than the N1 bonds, which may be due to the positive charge residing on N2: this was also observed in the structure of the nitrate salt (de Bonn et al., 2001 ▸). There are also several weak C—H⋯Br contacts (Table 1 ▸) in (I); the weak and strong interactions result in each bromide ion accepting a total of seven hydrogen bonds (Fig. 4 ▸).
(10) loops occur within the chain. The N2 bond is significantly shorter than the N1 bonds, which may be due to the positive charge residing on N2: this was also observed in the structure of the nitrate salt (de Bonn et al., 2001 ▸). There are also several weak C—H⋯Br contacts (Table 1 ▸) in (I); the weak and strong interactions result in each bromide ion accepting a total of seven hydrogen bonds (Fig. 4 ▸).
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1n⋯Br1i | 0.89 (2) | 2.68 (3) | 3.5666 (15) | 170.7 (18) |
| N1—H2n⋯Br1 | 0.89 (2) | 2.62 (2) | 3.5117 (14) | 175.0 (19) |
| N2—H3n⋯Br1ii | 0.87 (2) | 2.39 (2) | 3.2490 (13) | 173.3 (17) |
| C1—H1a⋯Br1i | 0.98 | 3.11 | 3.9690 (18) | 148 |
| C1—H1b⋯Br1iii | 0.98 | 3.09 | 4.0175 (19) | 158 |
| C1—H1c⋯Br1iv | 0.98 | 2.90 | 3.8682 (17) | 168 |
| C2—H2c⋯Br1iii | 0.98 | 3.07 | 3.9843 (18) | 156 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Figure 3.
Partial packing diagram for (I), showing the formation of [010] chains linked by N—H⋯Br hydrogen bonds. C-bound H atoms are omitted for clarity. Symmetry codes as in Table 1 ▸.
Figure 4.
The environment of the bromide ion in the crystal of (I). [Symmetry codes: (i)  − x,
− x,  − y,
− y,  − z; (ii)
− z; (ii)  − x,
− x,  − y,
− y,  − z; (iii) −x,
− z; (iii) −x,  + y,
+ y,  − y; (iv) x,
− y; (iv) x,  − y, z −
− y, z −  .] Note that each of the five cations has a different bonding mode: η1 N1, N2 and C1 and η2 N1 + C1 and C1 + C2.
.] Note that each of the five cations has a different bonding mode: η1 N1, N2 and C1 and η2 N1 + C1 and C1 + C2.
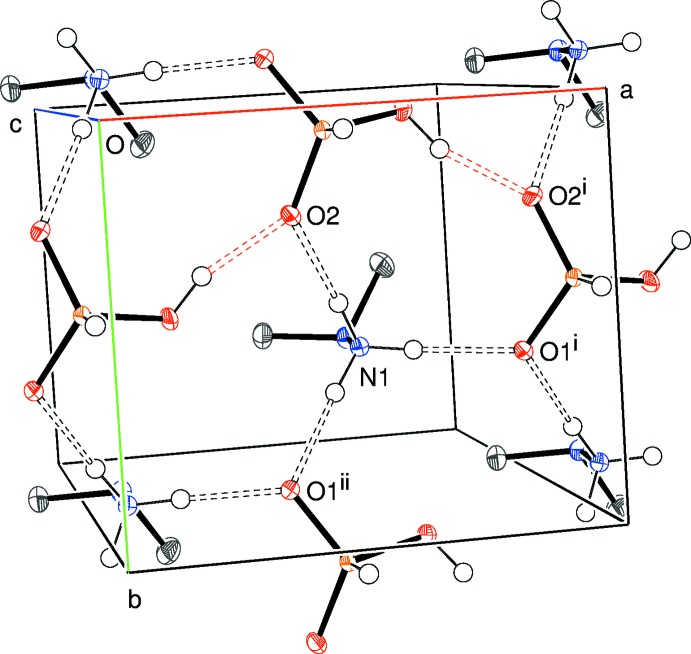
The crystal structure of (II) appears to correlate with the novel protonation pattern of the C2H9N2 + cation: the three H atoms attached to N1 each partake in a strong, near-linear N—H⋯O hydrogen bond to nearby H2PO3 − anions (Table 2 ▸). The anions are linked into [100] chains by O—H⋯O hydrogen bonds with adjacent anions in the chain related by a-glide symmetry. Together, these interactions generate (001) sheets (Fig. 5 ▸) As usual (Harrison, 2001 ▸), the P—H grouping of the anion does not participate in hydrogen bonds and the H atom points into the inter-layer region.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1n⋯O1i | 0.91 | 1.83 | 2.736 (2) | 176 |
| N1—H2n⋯O1ii | 0.91 | 1.85 | 2.762 (2) | 176 |
| N1—H3n⋯O2 | 0.91 | 1.91 | 2.814 (2) | 175 |
| O3—H1o⋯O2i | 0.87 | 1.74 | 2.568 (2) | 159 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 5.
Partial packing diagram for (II), showing part of an (001) sheet. Symmetry codes as in Table 2 ▸.
Database survey
A search of the Cambridge Structural Database (CSD; Groom et al., 2016 ▸) revealed the crystal structures of the four UDMH derivatives cited above: refcodes for the azide, nitrate, chloride and picrate salts are CORRUW, IBOLOA, FOHLUK and AZUXID, respectively.
Synthesis and crystallization
Caution! UDMH is toxic, potentially carcinogenic and may form explosive mixtures with oxidizing agents: all appropriate safety measures must be put in place when handling this compound.
To prepare (I), aqueous solutions of UDMH (10 ml, 1.0 M) and hydrobromic acid (10 ml, 1.0 M) were mixed at room temperature to yield a colourless solution and colourless rods (to ∼1 mm in length) of (I) grew as the solvent evaporated in a watch glass. These crystals are extremely hygroscopic and should be immediately transferred to a desiccator for storage: if left in air, they absorb enough water to completely dissolve within an hour or two.
To prepare (II), aqueous solutions of UDMH (10 ml, 1.0 M) and phosphorus acid (10 ml, 1.0 M) were mixed at room temperature to yield a colourless solution and yellowish slabs of (II) grew as the increasingly viscous solvent slowly evaporated over several days in a watch glass. These crystals are hygroscopic and should be stored in a desiccator. IR: 2383 cm−1 (P—H stretch).
The IR spectra of UDMH, (I) and (II) are available as supporting information.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The N-bound H atoms in (I) were located in difference maps and their positions freely refined; those in (II) were relocated to idealized locations and refined as riding atoms. The O-bound H atom in (II) was located in a difference map and refined as riding, in its as-found relative position. The methyl H atoms were geometrically placed (C—H = 0.98 Å): the –CH3 groups were allowed to rotate, but not to tip, to best fit the electron density. The constraint U iso(H) = 1.2U eq(carrier) or 1.5U eq(methyl carrier) was applied in all cases.
Table 3. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C2H9N2 +·Br− | C2H9N2 +·H2PO3 − |
| M r | 141.02 | 142.10 |
| Crystal system, space group | Monoclinic, I2/a | Orthorhombic, P n a21 |
| Temperature (K) | 100 | 100 |
| a, b, c (Å) | 13.2423 (2), 5.1239 (1), 16.1839 (3) | 8.0690 (2), 6.9970 (2), 11.7001 (6) |
| α, β, γ (°) | 90, 94.838 (2), 90 | 90, 90, 90 |
| V (Å3) | 1094.20 (3) | 660.57 (4) |
| Z | 8 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 7.36 | 0.35 |
| Crystal size (mm) | 0.23 × 0.09 × 0.09 | 0.18 × 0.18 × 0.02 |
| Data collection | ||
| Diffractometer | Rigaku Mercury CCD | Rigaku Mercury CCD |
| Absorption correction | Multi-scan (CrystalClear; Rigaku, 2012 ▸) | – |
| T min, T max | 0.282, 0.557 | – |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 6485, 1258, 1224 | 5347, 1395, 1365 |
| R int | 0.029 | 0.023 |
| (sin θ/λ)max (Å−1) | 0.649 | 0.649 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.020, 0.051, 1.12 | 0.025, 0.065, 1.09 |
| No. of reflections | 1258 | 1395 |
| No. of parameters | 58 | 77 |
| No. of restraints | 0 | 1 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.50, −0.48 | 0.24, −0.28 |
| Absolute structure | – | Refined as an inversion twin. |
| Absolute structure parameter | – | 0.15 (14) |
Supplementary Material
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989016011993/su5314sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016011993/su5314Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016011993/su5314IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989016011993/su5314Isup4.cml
Supporting information file. DOI: 10.1107/S2056989016011993/su5314IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the EPSRC National Crystallography Service (University of Southampton) for the data collections.
supplementary crystallographic information
(I) 1,1-Dimethylhydrazin-1-ium bromide. Crystal data
| C2H9N2+·Br− | F(000) = 560 |
| Mr = 141.02 | Dx = 1.712 Mg m−3 |
| Monoclinic, I2/a | Mo Kα radiation, λ = 0.71073 Å |
| a = 13.2423 (2) Å | Cell parameters from 5743 reflections |
| b = 5.1239 (1) Å | θ = 2.5–27.5° |
| c = 16.1839 (3) Å | µ = 7.36 mm−1 |
| β = 94.838 (2)° | T = 100 K |
| V = 1094.20 (3) Å3 | Rod, colourless |
| Z = 8 | 0.23 × 0.09 × 0.09 mm |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Data collection
| Rigaku Mercury CCD diffractometer | 1224 reflections with I > 2σ(I) |
| ω scans | Rint = 0.029 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2012) | θmax = 27.5°, θmin = 2.5° |
| Tmin = 0.282, Tmax = 0.557 | h = −17→17 |
| 6485 measured reflections | k = −6→5 |
| 1258 independent reflections | l = −20→19 |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.020 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.051 | w = 1/[σ2(Fo2) + (0.0349P)2 + 0.3874P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max = 0.001 |
| 1258 reflections | Δρmax = 0.50 e Å−3 |
| 58 parameters | Δρmin = −0.48 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2014 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0151 (6) |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.13506 (11) | 0.3029 (3) | 0.30499 (9) | 0.0197 (3) | |
| H1n | 0.1888 (17) | 0.204 (4) | 0.3204 (13) | 0.024* | |
| H2n | 0.1477 (17) | 0.393 (3) | 0.2599 (15) | 0.024* | |
| N2 | 0.13068 (10) | 0.4851 (2) | 0.37297 (8) | 0.0170 (3) | |
| H3n | 0.1867 (17) | 0.574 (4) | 0.3765 (12) | 0.020* | |
| C1 | 0.11750 (14) | 0.3372 (3) | 0.45000 (11) | 0.0191 (3) | |
| H1a | 0.1708 | 0.2047 | 0.4582 | 0.029* | |
| H1b | 0.0510 | 0.2519 | 0.4454 | 0.029* | |
| H1c | 0.1220 | 0.4571 | 0.4973 | 0.029* | |
| C2 | 0.04544 (15) | 0.6693 (3) | 0.35325 (15) | 0.0278 (4) | |
| H2a | 0.0567 | 0.7656 | 0.3025 | 0.042* | |
| H2b | 0.0416 | 0.7925 | 0.3992 | 0.042* | |
| H2c | −0.0182 | 0.5715 | 0.3450 | 0.042* | |
| Br1 | 0.17057 (2) | 0.64136 (3) | 0.12100 (2) | 0.01501 (11) |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0198 (7) | 0.0244 (6) | 0.0150 (7) | 0.0014 (6) | 0.0030 (5) | 0.0027 (5) |
| N2 | 0.0120 (6) | 0.0147 (6) | 0.0240 (7) | −0.0019 (5) | −0.0004 (5) | 0.0017 (5) |
| C1 | 0.0195 (8) | 0.0226 (8) | 0.0151 (8) | −0.0015 (5) | 0.0010 (6) | −0.0008 (5) |
| C2 | 0.0175 (9) | 0.0173 (8) | 0.0479 (12) | 0.0030 (6) | −0.0014 (8) | 0.0067 (7) |
| Br1 | 0.01199 (13) | 0.01558 (14) | 0.01745 (14) | −0.00069 (4) | 0.00116 (7) | 0.00263 (4) |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Geometric parameters (Å, º)
| N1—N2 | 1.4478 (19) | C1—H1a | 0.98 |
| N1—H1n | 0.89 (2) | C1—H1b | 0.98 |
| N1—H2n | 0.89 (2) | C1—H1c | 0.98 |
| N2—C1 | 1.482 (2) | C2—H2a | 0.98 |
| N2—C2 | 1.485 (2) | C2—H2b | 0.98 |
| N2—H3n | 0.87 (2) | C2—H2c | 0.98 |
| N2—N1—H1n | 103.6 (14) | H1a—C1—H1b | 109.5 |
| N2—N1—H2n | 108.0 (12) | N2—C1—H1c | 109.5 |
| H1n—N1—H2n | 108.8 (19) | H1a—C1—H1c | 109.5 |
| N1—N2—C1 | 108.93 (12) | H1b—C1—H1c | 109.5 |
| N1—N2—C2 | 108.97 (14) | N2—C2—H2a | 109.5 |
| C1—N2—C2 | 111.38 (14) | N2—C2—H2b | 109.5 |
| N1—N2—H3n | 107.4 (13) | H2a—C2—H2b | 109.5 |
| C1—N2—H3n | 111.9 (13) | N2—C2—H2c | 109.5 |
| C2—N2—H3n | 108.1 (13) | H2a—C2—H2c | 109.5 |
| N2—C1—H1a | 109.5 | H2b—C2—H2c | 109.5 |
| N2—C1—H1b | 109.5 |
(I) 1,1-Dimethylhydrazin-1-ium bromide. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1n···Br1i | 0.89 (2) | 2.68 (3) | 3.5666 (15) | 170.7 (18) |
| N1—H2n···Br1 | 0.89 (2) | 2.62 (2) | 3.5117 (14) | 175.0 (19) |
| N2—H3n···Br1ii | 0.87 (2) | 2.39 (2) | 3.2490 (13) | 173.3 (17) |
| C1—H1a···Br1i | 0.98 | 3.11 | 3.9690 (18) | 148 |
| C1—H1b···Br1iii | 0.98 | 3.09 | 4.0175 (19) | 158 |
| C1—H1c···Br1iv | 0.98 | 2.90 | 3.8682 (17) | 168 |
| C2—H2c···Br1iii | 0.98 | 3.07 | 3.9843 (18) | 156 |
Symmetry codes: (i) −x+1/2, −y+1/2, −z+1/2; (ii) −x+1/2, −y+3/2, −z+1/2; (iii) −x, y−1/2, −z+1/2; (iv) x, −y+3/2, z+1/2.
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Crystal data
| C2H9N2+·H2PO3− | Dx = 1.429 Mg m−3 |
| Mr = 142.10 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 4031 reflections |
| a = 8.0690 (2) Å | θ = 3.4–27.5° |
| b = 6.9970 (2) Å | µ = 0.35 mm−1 |
| c = 11.7001 (6) Å | T = 100 K |
| V = 660.57 (4) Å3 | Plate, yellow |
| Z = 4 | 0.18 × 0.18 × 0.02 mm |
| F(000) = 304 |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Data collection
| Rigaku Mercury CCD diffractometer | Rint = 0.023 |
| ω scans | θmax = 27.5°, θmin = 3.4° |
| 5347 measured reflections | h = −8→10 |
| 1395 independent reflections | k = −8→9 |
| 1365 reflections with I > 2σ(I) | l = −15→13 |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.025 | H-atom parameters constrained |
| wR(F2) = 0.065 | w = 1/[σ2(Fo2) + (0.0374P)2 + 0.203P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.09 | (Δ/σ)max < 0.001 |
| 1395 reflections | Δρmax = 0.24 e Å−3 |
| 77 parameters | Δρmin = −0.28 e Å−3 |
| 1 restraint | Absolute structure: Refined as an inversion twin. |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.15 (14) |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refined as a 2-component inversion twin. |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.5416 (2) | 0.5852 (2) | 0.22552 (17) | 0.0132 (4) | |
| H1n | 0.6455 | 0.5972 | 0.1962 | 0.016* | |
| H2n | 0.4779 | 0.6848 | 0.2016 | 0.016* | |
| H3n | 0.4960 | 0.4735 | 0.2011 | 0.016* | |
| N2 | 0.5499 (2) | 0.5853 (3) | 0.34964 (17) | 0.0145 (4) | |
| C1 | 0.3807 (3) | 0.5724 (3) | 0.3936 (3) | 0.0189 (5) | |
| H1a | 0.3169 | 0.6834 | 0.3677 | 0.028* | |
| H1b | 0.3832 | 0.5699 | 0.4773 | 0.028* | |
| H1c | 0.3286 | 0.4551 | 0.3652 | 0.028* | |
| C2 | 0.6471 (3) | 0.4186 (3) | 0.3845 (2) | 0.0206 (5) | |
| H2a | 0.7575 | 0.4255 | 0.3499 | 0.031* | |
| H2b | 0.5912 | 0.3018 | 0.3590 | 0.031* | |
| H2c | 0.6576 | 0.4169 | 0.4679 | 0.031* | |
| P1 | 0.45797 (6) | 0.05934 (7) | 0.10615 (6) | 0.01209 (15) | |
| H1 | 0.4666 | 0.0529 | −0.0064 | 0.015* | |
| O1 | 0.35870 (18) | −0.1101 (2) | 0.14409 (14) | 0.0153 (3) | |
| O2 | 0.39202 (17) | 0.2526 (2) | 0.13797 (14) | 0.0165 (4) | |
| O3 | 0.63746 (19) | 0.0285 (2) | 0.15307 (17) | 0.0196 (4) | |
| H1o | 0.7059 | 0.1224 | 0.1417 | 0.024* |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0099 (8) | 0.0108 (8) | 0.0189 (10) | 0.0002 (6) | −0.0001 (7) | −0.0003 (7) |
| N2 | 0.0114 (9) | 0.0142 (9) | 0.0180 (11) | 0.0005 (6) | −0.0007 (7) | −0.0007 (8) |
| C1 | 0.0124 (10) | 0.0206 (11) | 0.0236 (13) | −0.0007 (8) | 0.0021 (10) | −0.0016 (9) |
| C2 | 0.0172 (11) | 0.0209 (12) | 0.0237 (13) | 0.0050 (8) | −0.0028 (10) | 0.0023 (10) |
| P1 | 0.0074 (2) | 0.0095 (2) | 0.0194 (3) | 0.00031 (18) | 0.0003 (3) | 0.0004 (2) |
| O1 | 0.0097 (6) | 0.0100 (7) | 0.0263 (9) | −0.0005 (6) | 0.0011 (6) | 0.0013 (6) |
| O2 | 0.0097 (6) | 0.0112 (7) | 0.0286 (10) | 0.0017 (6) | −0.0011 (6) | −0.0023 (6) |
| O3 | 0.0075 (6) | 0.0127 (7) | 0.0386 (10) | −0.0009 (6) | −0.0030 (7) | 0.0040 (7) |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Geometric parameters (Å, º)
| N1—N2 | 1.454 (3) | C2—H2a | 0.98 |
| N1—H1n | 0.91 | C2—H2b | 0.98 |
| N1—H2n | 0.91 | C2—H2c | 0.98 |
| N1—H3n | 0.91 | P1—O1 | 1.4982 (15) |
| N2—C1 | 1.462 (3) | P1—O2 | 1.5003 (16) |
| N2—C2 | 1.463 (3) | P1—O3 | 1.5638 (16) |
| C1—H1a | 0.98 | P1—H1 | 1.32 |
| C1—H1b | 0.98 | O3—H1o | 0.8689 |
| C1—H1c | 0.98 | ||
| N2—N1—H1n | 109.5 | H1b—C1—H1c | 109.5 |
| N2—N1—H2n | 109.5 | N2—C2—H2a | 109.5 |
| H1n—N1—H2n | 109.5 | N2—C2—H2b | 109.5 |
| N2—N1—H3n | 109.5 | H2a—C2—H2b | 109.5 |
| H1n—N1—H3n | 109.5 | N2—C2—H2c | 109.5 |
| H2n—N1—H3n | 109.5 | H2a—C2—H2c | 109.5 |
| N1—N2—C1 | 107.94 (18) | H2b—C2—H2c | 109.5 |
| N1—N2—C2 | 107.62 (17) | O1—P1—O2 | 116.76 (9) |
| C1—N2—C2 | 110.69 (18) | O1—P1—O3 | 106.37 (9) |
| N2—C1—H1a | 109.5 | O2—P1—O3 | 111.46 (9) |
| N2—C1—H1b | 109.5 | O1—P1—H1 | 107.3 |
| H1a—C1—H1b | 109.5 | O2—P1—H1 | 107.3 |
| N2—C1—H1c | 109.5 | O3—P1—H1 | 107.3 |
| H1a—C1—H1c | 109.5 | P1—O3—H1o | 115.5 |
(II) 2,2-Dimethylhydrazin-1-ium dihydrogen phosphite. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1n···O1i | 0.91 | 1.83 | 2.736 (2) | 176 |
| N1—H2n···O1ii | 0.91 | 1.85 | 2.762 (2) | 176 |
| N1—H3n···O2 | 0.91 | 1.91 | 2.814 (2) | 175 |
| O3—H1o···O2i | 0.87 | 1.74 | 2.568 (2) | 159 |
Symmetry codes: (i) x+1/2, −y+1/2, z; (ii) x, y+1, z.
References
- Bonn, O. de, Hammerl, A., Klapötke, T. M., Mayer, P., Piotrowski, H. & Zewen, H. (2001). Z. Anorg. Allg. Chem. 627, 2011–2015.
- Edwards, T. (2003). J. Propul. Power, 19, 1089–1107.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Harrison, W. T. A. (2001). J. Solid State Chem. 160, 4–7.
- Harrison, W. T. A. (2003). Acta Cryst. E59, o1351–o1353.
- Klapötke, T. M., Nöth, H., Schwenk-Kircher, H., Walther, W. H. & Holl, G. (1999). Polyhedron, 18, 717–719.
- Merkoulov, A., Harms, K. & Sundermeyer, J. (2005). Acta Cryst. E61, o1800–o1801.
- Mu, X.-G., Wang, X.-J., Liu, X.-X., Cui, H. & Wang, H. (2011). Acta Cryst. E67, o2749. [DOI] [PMC free article] [PubMed]
- Rigaku (2012). CrystalClear. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S2056989016011993/su5314sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016011993/su5314Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016011993/su5314IIsup3.hkl
Supporting information file. DOI: 10.1107/S2056989016011993/su5314Isup4.cml
Supporting information file. DOI: 10.1107/S2056989016011993/su5314IIsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report