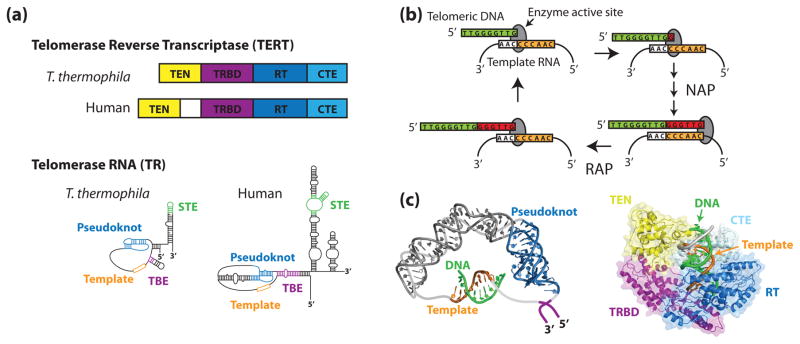
Figure 1. The telomerase ribonucleoprotein complex.
(a) Tetrahymena thermophila and human TERTs and TRs share a similar domain organization. In both species, TERT is composed of an N-terminal (TEN) domain, an RNA-binding domain (TRBD), a reverse transcriptase domain (RT), and a C-terminal extension (CTE). Conserved RNA structural features include an RNA pseudoknot fold, template, template boundary element (TBE), and a stem terminal element (STE). (b) During the telomerase catalytic cycle, telomeric DNA is positioned in the enzyme active site by basepairing alignment with the template RNA. Individual nucleotides are reverse transcribed off of the template RNA in a process known as nucleotide addition processivity (NAP). When the end of the template is reached, the DNA-RNA duplex is denatured and the RNA is re-positioned to add another repeat, a process known as repeat addition processivity (RAP). (c) In silico modeling of human TR (left) and human TERT (right) (adapted from [1,2] with permission). Zhang and co-workers used residual dipolar coupling orientation constraints to model the core region of human TR using existing NMR structures. Steczkiewicz et al. used homology modeling and structure prediction algorithms to model human TERT based on solved structures of the Tribolium castaneum TERT and the Tetrahymena thermophila TEN and TRBD domains, and used molecular docking to predict the position of the TEN domain within the enzyme.