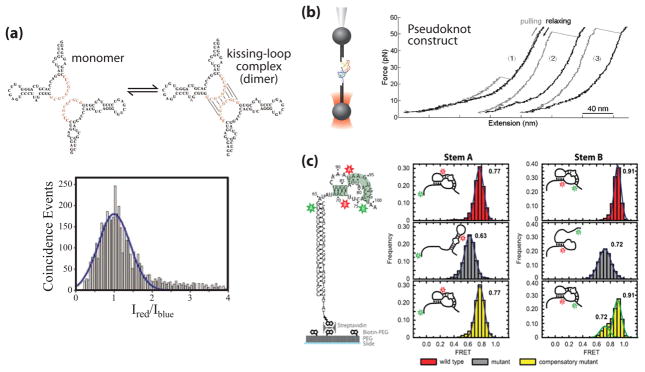
Figure 3. Single molecule studies of telomerase RNA structure.
(a) Proposed dimerization interaction in the J7b/8a region of the CR7 region of hTR (adapted from [3] with permission). RNA is 5′ end labeled with either Alexa 488 or Alexa 647 in a 1:1 ratio and diluted to a total concentration of 100 pM. A histogram of the log ratio of red to blue fluorescence for single molecules diffusing through the excitation volume was indicative of a single distribution centered at a 1:1 ratio, corresponding to the hTR-Alexa 488: hTR-Alexa 647 dimer. (b) Mechanical folding/unfolding of the hTR pseudoknot domain RNA (adapted from [4] with permission). The pseudoknot construct used in this study was oriented between two DNA handles and held in an optical trap. The RNA molecule is pulled (grey line) and relaxed (black line) at a constant rate of 100 nm/sec as the resultant force is measured. The rip at ~24 pN (trace 1) corresponds to the unfolding force of an alternative hairpin conformation observed in a subset of traces. The rips at ~50 pN (traces 2 & 3) correspond to the unfolding force of the complete pseudoknot, resulting in a rip size of ~36 nm which corresponds to the dimensions of the pseudoknot. (c) Single molecule FRET (smFRET) studies of the Tetrahymena TR (tTR) pseudoknot domain RNA (adapted from [5] with permission). Constructs were labeled with a donor dye (green) at residue U63 and an acceptor dye (red) at U92 to measure folding of stem A or labeled with a donor dye at U73 and an acceptor dye at U99 to measure folding of stem B. smFRET histograms were obtained with the stem A and stem B labeling sites for the wild-type RNA sequence (red), a mutant pseudoknot designed to disrupt basepairing in stem A (grey), and a compensatory mutant designed to restore basepairing (yellow).