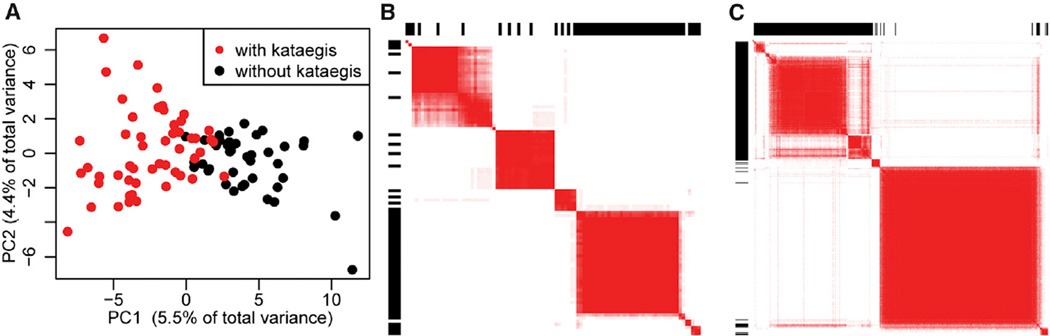
Figure 4. Transcriptome-wide Kataegis Expression Signature.
(A) PCA analysis of the levels of 628 differentially expressed genes in the 97 tumors separates samples with (red) and without (black) kataegis into two classes.
(B) Heatmap showing results of consensus clustering using the first five principal components in the 97 tumors. Samples with kataegis are indicated by a black bar in the outer rectangle area. The intensity of the heatmap represents the number of times (over a total of 20 runs) each pair of samples is clustered together (white = 0, red = 20).
(C) Heatmap showing consensus clusters using the first five principal components in 412 tumors. Samples predicted to harbor kataegis loci by the GLM (95% CI) are indicated by a black bar in the outer rectangle area.