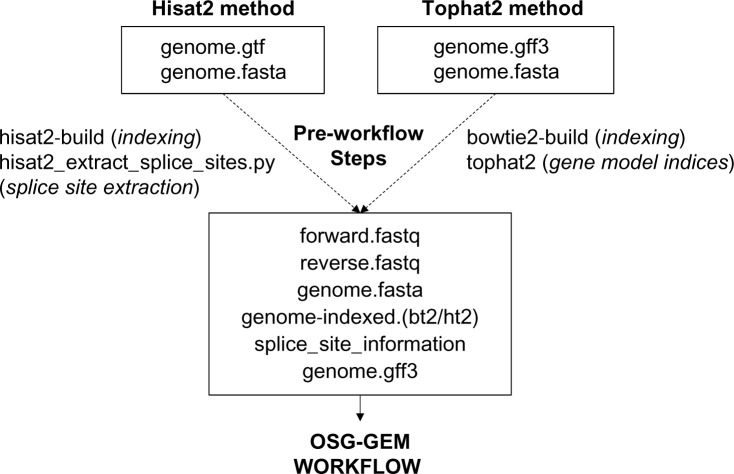
Figure 2.
Preparation of input files for the gene expression matrix construction workflow on the Open Science Grid (OSG-GEM). Required input files for either the Hisat2 or the Tophat2 method are shown in boxes. The user provides paired-end DNA sequences in the FASTQ format (forward/reverse), which can be extracted from SRA format files with the NCBI SRA toolkit. The reference genome (genome) in the FASTA format must be indexed using either the Hisat2 or the Bowtie2 application. Built into the Hisat2 software package, the hisat2_extract_splice_sites.py script can generate a tab-delimited list of splice sites using a reference annotation file in the GTF format. Tophat2 can generate a set of gene model indices from GFF3 or GTF format files that contain splice site information in the form of a reference transcriptome. FASTQ file locations are defined in the osg-gem.config file and all other files are placed in the reference directory of the OSG-GEM workflow.