Figure 3.
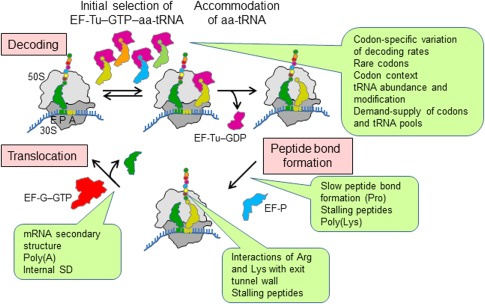
Links between translation elongation and non‐uniform translation. Elongation entails three steps, decoding, peptide bond formation and translocation. During decoding, EF‐Tu in bacteria (or eEF1α in eukaryotes) delivers aa‐tRNA to the A site of the ribosome. These factors are GTPases that in their GTP‐bound conformation form a high‐affinity ternary complex with aa‐tRNA and GTP which, in turn, binds to the ribosome and, after GTP hydrolysis, releases aa‐tRNA to accommodate in the PTC. The ribosome selects an aa‐tRNA that is cognate to the codon in the A site (yellow) among other aa‐tRNAs. These can be almost‐cognate (orange), near‐cognate (green) or non‐cognate (blue). Peptide bond formation between aa‐tRNA in the A site and pept‐tRNA in the P site is catalyzed by the ribosome and usually does not require auxiliary proteins. The peptidyl transfer reaction between two Pro residues requires EF‐P (eIF5A in eukaryotes). Translocation is catalyzed by EF‐G (eEF2 in eukaryotes) at the cost of GTP hydrolysis. Ribosomes, aa‐tRNA and factors are all active players, and the exact interplay between them determines the actual speed and fidelity of translation. Callouts summarize potential sources of translational non‐uniformity at each step of the elongation cycle.
