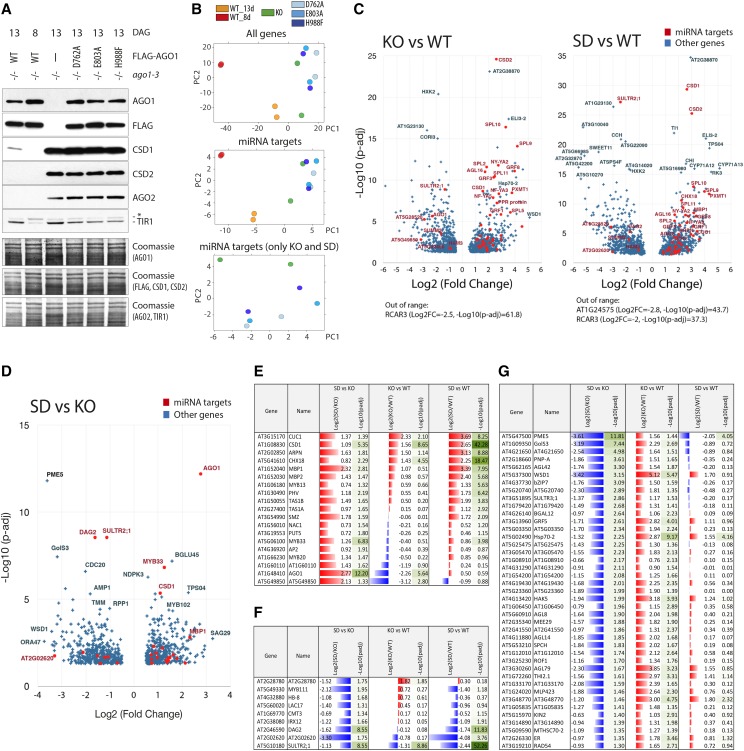
Figure 3.
AGO1 slicer-deficient alleles do not provide evidence for slicer-independent miRNA target regulation. A, Western-blot analysis of protein extracts from 8- or 13-d old whole seedlings of ago1-3 and stable transgenic lines expressing the wild type or slicer-deficient FLAG-AGO1 (D762A, E803A, and H988F) in the ago1-3 null background. Three identical membranes were cut horizontally according to the expected protein sizes and were used for incubation with antibodies detecting the miRNA targets CSD1 (miR398), CSD2 (miR398), AGO2 (miR403), and TIR1 (miR393). AGO1 and FLAG antibodies were used to monitor expression of FLAG-AGO1 in the transgenic lines. Coomassie staining serves as loading control. B to G, Analysis of RNA-seq data from poly(A+) RNA isolated from seedlings described in A. Two biological replicates of each genetic background were analyzed. B, PCA. KO, ago1-3 null (knockout); SD, ago1-3 expressing slicer-deficient AGO1 mutants. C and D, Volcano plot showing differential expression between 13-d-old seedlings of ago1-3 (KO) compared to ago1-3 FLAG-AGO1 (WT) and averaged data from slicer-deficient samples (SD) compared to the wild type (C) or SD compared to KO (D). Only significantly regulated genes are plotted (|log2(fold change)| > 0.5, adjusted P value < 0.05). Validated or predicted miRNA targets are highlighted in red. E, miRNA targets upregulated in SD compared to KO. F, miRNA targets downregulated in SD compared to KO. G, Genes significantly downregulated in SD compared to KO and significantly upregulated in KO compared to wild-type plants. In E to G, only genes with |log2(fold change)| > 0.5, adjusted P value < 0.05 are included. Adjusted P values were calculated using Wald test with Benjamini-Hochberg adjustment in all cases.