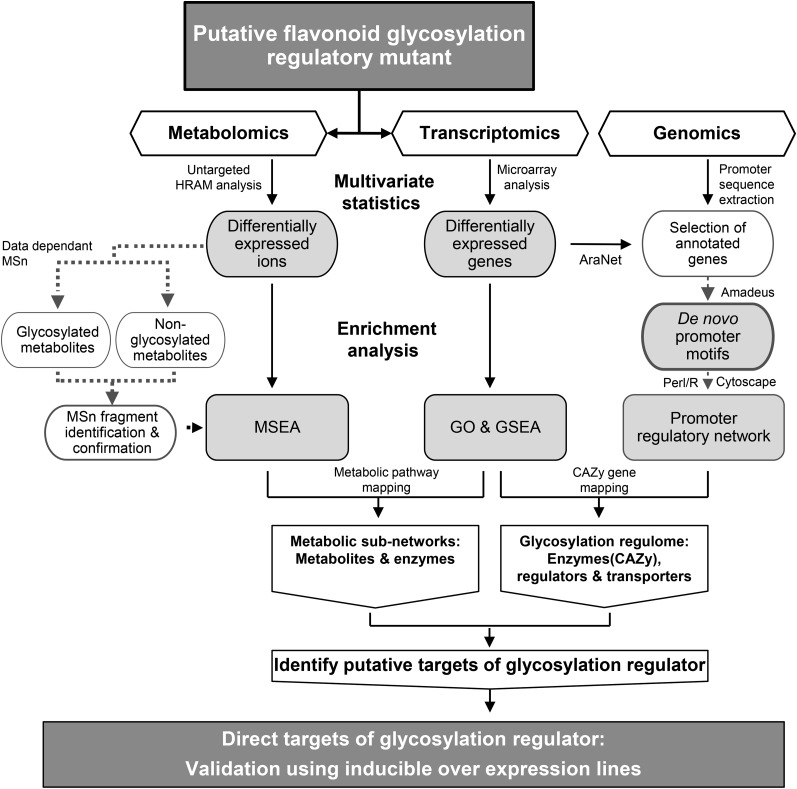
Figure 1.
Integrative omics strategy to identify metabolic targets and regulome of a flavonoid glycosylation regulator. Differential metabolites and transcripts were identified from untargeted metabolite and expression profiling of a putative glycosylation regulatory mutant, tt8 (Ws). Glycosylated metabolites affected in tt8 (Ws) were identified by mass fragmentation. Integration workflow consists of two stages: (1) mapping enriched metabolites and gene sets onto metabolic pathways; and (2) mapping of enriched genes onto regulatory network based on promoter motif similarity between differentially expressed genes. The metabolites and enzymes of the metabolic subnetworks obtained from the first stage are further mapped onto the glycosylation regulome from the second stage. Finally, the overlapping putative targets of the regulator are validated using inducible overexpression (in Col background) and mutant lines (in Ws background) based on their reciprocal patterns of accumulation. GO, Gene ontology; GSEA, gene set enrichment analysis; MSEA, metabolite set enrichment analysis; HRAM, high-resolution accurate mass.