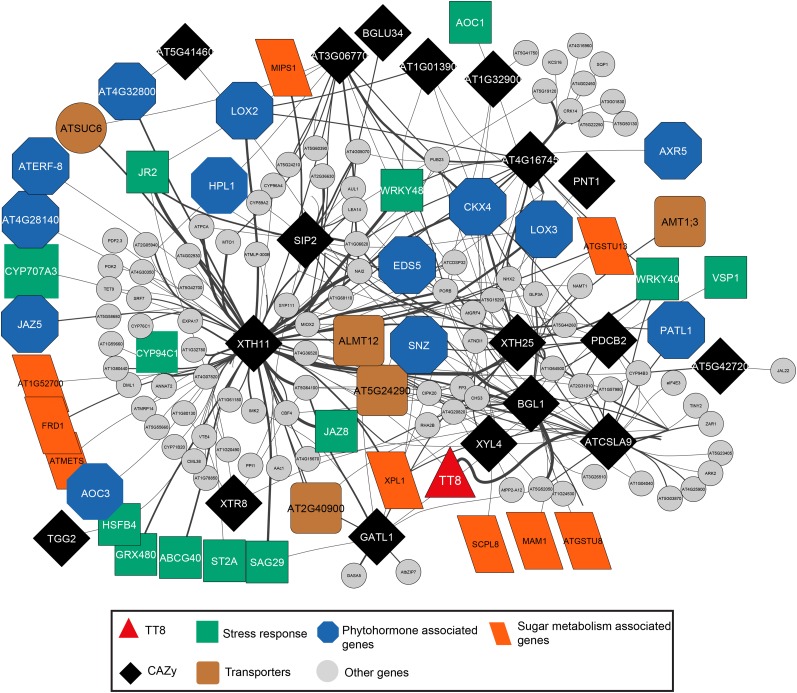
Figure 4.
CAZy coding genes share motif similarity with stress response and phytohormone-associated genes. Differentially expressed genes form the nodes, while the edges are based on sharing of promoter motifs. To identify relationships between sugar metabolism genes and those of other pathways in the regulome, a constraint was applied to connect nodes of sugar metabolism genes that share a minimum of 14 motifs (top 25 percentile) with other genes. This constrained regulatory network is shown here. The width of the edges corresponds to the number of shared motifs between the two nodes, with high similarity resulting in thicker edges. This glycosylation regulome consists of 18 enzymes (CAZy) connected mostly with stress response genes (13 genes) and phytohormone-associated genes (13 genes), followed by transporters (five genes) and and transferases, hydrolases, and oxidases (nine genes). Other genes in the glycosylation network but not directly annotated to any of the above functional processes are shown as gray circles.