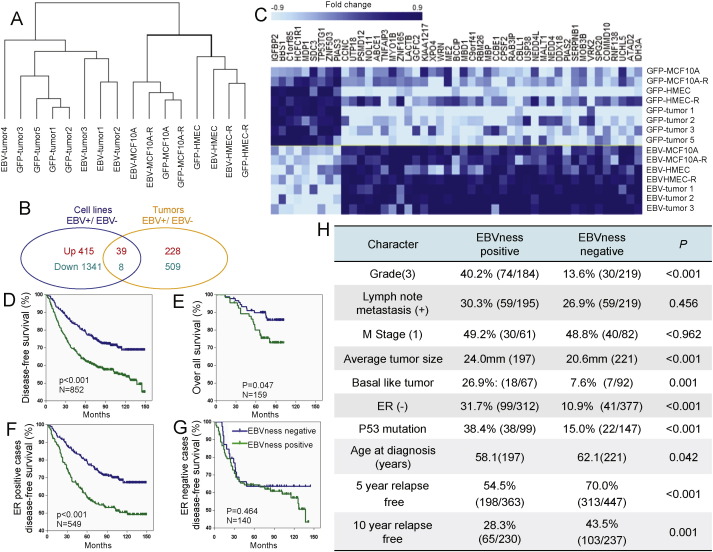
Fig. 6.
An EBVness-signature for breast cancer correlates with poor clinical out come. (A) Cluster analysis of RNA expression profiles from 16 samples, including 4 EBV-infected MEC lines, 4 GFP-control cell lines, 4 EBV-MEC-derived breast cancers and 4 GFP-control tumors. EBV-tumor 4 was an outlier and is clustered singly, and was therefore not included in the following analysis. (B) Genes that were significantly different (P ≤ 0.001) between EBV-infected and GFP-control MECs (blue circle) and between EBV-infected BC and GFP-control BC (yellow circle) were determined separately. Differentially regulated genes were identified (39 up-regulated, and 8 down-regulated) as candidates for “EBVness” signature training. (C) A heatmap was generated displaying the 47 genes significantly and differentially expressed in EBV-infected samples both in vitro and in vivo. For further details see Table S1. (D–G) The “EBVness” signature is associated with decreased disease-free and overall survival. The identified gene signature was reduced from 47 to 37 to allow for analysis of archival datasets (HG-U133A chip) published in GEO. The data represent a pooled analysis of four data sets including GSE1456, GSE2990 GSE4922 and GSE2034 (HG-U133A platform). For further details and separate analysis see Fig. S4. (F) Clinicopathologic characteristics of breast cancers that carry the “EBVness” feature, again based on a pooled analysis of four data sets including GSE1456, GSE2990 GSE4922 and GSE2034 (HG-U133A platform). Also see Table S5.