Abstract
Autism spectrum disorders affect millions of individuals worldwide, but their heterogeneity complicates therapeutic intervention that is essentially symptomatic. A versatile yet relevant model to rationally screen among hundreds of therapeutic options would help improving clinical practice. Here we investigated whether neurons differentiated from pluripotent stem cells can provide such a tool using SHANK3 haploinsufficiency as a proof of principle. A library of compounds was screened for potential to increase SHANK3 mRNA content in neurons differentiated from control human embryonic stem cells. Using induced pluripotent stem cell technology, active compounds were then evaluated for efficacy in correcting dysfunctional networks of neurons differentiated from individuals with deleterious point mutations of SHANK3. Among 202 compounds tested, lithium and valproic acid showed the best efficacy at corrected SHANK3 haploinsufficiency associated phenotypes in cellulo. Lithium pharmacotherapy was subsequently provided to one patient and, after one year, an encouraging decrease in autism severity was observed. This demonstrated that pluripotent stem cell-derived neurons provide a novel cellular paradigm exploitable in the search for specific disease-modifying treatments.
Abbreviations: ADHD, Attention Deficit Hyperactive Disorder; ADOS, Autism Diagnosis Observational Scale; ASD, autism spectrum disorders; DMSO, dimethylsulfoxyde; FDA, Food and Drug Administration; GAS, Global Assessment Scale; HTS, high throughput screening; hESC, human embryonic stem cells; iPSC, induced pluripotent stem cells; NSC, neural stem cells; PPIA, Peptidylprolyl Isomerase A; PPVT, Peabody Picture Vocabulary Test; PSC, pluripotent stem cells; RPM, Raven's progressive matrices; SHANK3, SH3 and multiple ankyrin repeat domains 3; SRS, Social Responsiveness Scale; VPA, valproic acid
Keywords: Autism, SHANK3, High throughput screening, Valproate, Lithium, Drug repurposing
Highlights
-
•
Human neurons were used to screen for compounds correcting symptoms associated with SHANK3 haploinsufficiency syndrome.
-
•
Screening criteria were the ability to increase SHANK3 expression and to increase glutamatergic transmission.
-
•
Selected hit compounds were then validated using neurons differentiated from individuals with SHANK3 disrupting mutations.
-
•
Lithium was selected and delivered to one of SHANK3 patient showing encouraging positive clinical outcomes after one year.
The clinical heterogeneity between individuals affected by autism makes it difficult to anticipate the effectiveness of a treatment. Furthermore, clinical practice lacks biological tools to help make such decisions. Here we use neurons, produced from pluripotent stem cells derived from patients affected by SHANK3 haploinsufficiency syndrome, to test the efficiency of therapeutic compounds. We screened the biological activity of more than 200 compounds on SHANK3 expression. Lithium was ultimately selected and delivered to one patient with a SHANK3-disruptive mutation. This resulted in a positive outcome, as determined by improved autistic core symptoms, thus supporting the usefulness of this type of predictive approach.
1. Introduction
Autism spectrum disorders (ASD) are a heterogeneous group of neurodevelopmental disorders that affect 1% of the population in developed countries and are characterized by persistent impairment in reciprocal social communication and restricted repetitive patterns of behavior (Newschaffer et al., 2007). Challenges in developing efficient pharmacotherapies for ASD reside in the limited understanding of ASD etiology, the difficulties to experimentally model the disease, the heterogeneity of symptoms and the large variety of potential triggers. Nonetheless, in around a third of patients a genetic cause has been identified (Kumar and Christian, 2009) and there appears to be a convergence on mechanisms controlling synaptic homeostasis and neuronal connectivity (Kumar and Christian, 2009, Toro et al., 2010, Bourgeron, 2015). Synapses and altered neuronal networks could be a relevant target to develop new therapeutic strategies for ASD (Spooren et al., 2012, Delorme et al., 2013). Improved understanding of the molecular substrates of autism is raising the hope of innovative “disease-modifying” treatments for altering the course of the disorder.
However, while resulting in similar pattern core of symptoms, the synaptic/connectivity alterations in individuals with ASD show a considerable degree of heterogeneity, which may mask the potential beneficial effects of a tested treatment if it is not addressed properly to a subgroup of individuals stratified on the basis of the affected genes or pathways rather than on the clinical phenotype per se (Delorme et al., 2013). This pleads for the design of new experimental and pre-clinical models to identify more personalized pharmacotherapies, approaches that require an access to relevant cellular models and tools that would allow analyzing at high throughput the effects of potential therapeutic compounds on human neurons.
In this light, we have previously shown that glutamatergic cortical neurons can be specifically differentiated from pluripotent stem cells (PSC) of either embryonic origin (human embryonic stem cells, hESC) or reprogrammed from somatic cells of patients with ASD caused by SHANK3 haplo-insufficiency (human induced pluripotent stem cells, hiPSC), and that they can serve as a cellular paradigm for high throughput screening (HTS) campaigns (Boissart et al., 2013). Permitting the systematic comparison of hundreds of small molecules in parallel, this can therefore accelerate identification of therapeutic compounds targeting pathophysiological mechanisms underlying distinct forms of ASD. The goal of the present study was to verify such a strategy by focusing on SHANK3 a key autism risk gene that codes for a synaptic scaffolding synaptic protein.
The synaptic protein SHANK3 has been linked to autism in different genetic screens where deleterious mutations affecting the SHANK3 gene were present in 0.69 to 2.12% of individuals (Durand et al., 2007, Leblond et al., 2014). SHANK3 is an abundant component of the post-synaptic density (PSD) where it acts as a scaffolding protein recruiting key post-synaptic elements, such as glutamate receptors, and linking them to the actin cytoskeleton (Boeckers et al., 2002). This scaffolding role contributes to the formation, stabilization and strengthening of the glutamatergic synapses and increases the efficiency of the glutamatergic transmission (Grabrucker et al., 2011). SHANK3 Knock-Out mice recapitulate some key autistic behaviors and show reduced number and activity of glutamatergic synapses as well as loss in experience-linked plasticity (Peca et al., 2011). At the cellular level, neurons differentiated from iPSC derived from individuals with SHANK3 haploinsufficiency, exhibit impaired electrophysiological responses to glutamatergic synapse stimulations that can be corrected by re-introducing SHANK3 cDNA expression (Shcheglovitov et al., 2013). Decreased glutamatergic neurotransmission thus appears as a central feature of SHANK3 haploinsufficiency and it can be reversed by restoring correct dosage of SHANK3 even during adulthood (Toro et al., 2010). In individuals with ASD, de novo loss-of-function mutations of SHANK3 reported in the literature affect only one allele of the gene, offering the opportunity to enhance transcription of the second allele, or improve mRNA stability and translation. We have therefore developed a high throughput molecular screening assay in order to pinpoint compounds increasing SHANK3 mRNA levels in human neurons derived from pluripotent stem cells.
2. Methods
2.1. Human Pluripotent Stem Cells (hPSC) Origin
One hESC and 4 iPSC lines were used in this study. The male hESC line SA001 was obtained from Cellartis (Goteborg, Sweden). The work involving hESC line was supervised by the French Bioethics Agency (Agreement number NOR AFSB 12002 43S). The control iPSC line 1869 was published previously (Boissart et al., 2013) and reprogrammed from fibroblasts obtained from the Coriell's biorepository (Coriell Institute for Medical Research, NJ, USA). The control line PB12 was reprogrammed from PBMC obtained from an anonymous blood donor at the French Blood Donor Organization. SHANK3-iPSC lines were derived from fibroblasts of 2 children with ASD carrying independent de novo SHANK-3 mutations. The 2 patients were diagnosed with autism and severe intellectual deficiency at the Robert Debré and Pitié-Salpétrière Hospitals according to DSM-IVTR criteria. Patient 1 (AUN-003, Leblond et al., 2014) carries a de novo truncating mutation P.Glu809X in exon 21 and corresponds to iPSC line SHANK3-STOP. Patient 2 carries a frame shift mutation (c.3853_3857dup, p. Gly1287Alafs X15) and corresponds to the iPSC line SHANK3 frame-shift. Both were included initially in a national observational study (IDRCB 2008-A00019-46). After patient's legal representatives approval, 8-mm skin punch biopsies were obtained (study approval by Committee for the Protection of Persons, CPP no. C07-33). Fibroblasts were derived from the donated tissue and reprogrammed using the four human genes OCT4, SOX2, c-Myc, and KLF4 cloned in Sendai viruses (Life Technologies, Carlsbad, CA, USA) and iPSC lines characterized according to Nakagawa et al. (2008). The same procedure was used to reprogrammed and characterized control iPSC lines.
2.2. Derivation of Cortical Neural Stem Cells (NSC) from PSC
Commitment of PSC to the neural lineage and derivation of stable cortical NSC was described previously (Boissart et al., 2012, Boissart et al., 2013). Briefly, neural commitment was induced using the BMP inhibitor Noggin (500 ng/ml, Peprotech, London, UK) and the Nodal inhibitor SB431542 (20 μM, Tocris Biosciences, Ellisville, Missouri, USA). At day 10, neural rosettes containing neuro-epithelial cells were collected and plated en bloc in poly-l-ornithin/laminin treated culture dishes in N2B27 medium containing Epidermal Growth Factor (EGF, 10 ng/ml, Peprotech, Neuilly, France), FGF-2 (10 ng/ml, Peprotech) and Brain-derived Growth Factor (BDNF, 20 ng/ml, Peprotech). At confluence, the passages were performed using trypsin at a density of 50,000 cells/cm2. Mass amplification was performed until passage 8 and cells were frozen. To start the terminal differentiation as post-mitotic neurons, cortical NSC were dissociated and plated in N2B27 without growth factors.
2.3. Compound Library
Two hundred and two small molecules were investigated, belonging to two commercially available libraries (Screen-Well® Kinase Inhibitor library, Enzo-life Sciences, 76 compounds and Screen-Well® Epigenetics library, Enzo-life Sciences, 43 compounds), I-stem laboratory reference compounds (Boissart et al., 2013, 50 compounds) and FDA approved drugs or related active principle (33 compounds). The complete list of compounds is provided in Table S1.
2.4. Automated and High Throughput Quantification of SHANK3 mRNA in Human Neurons
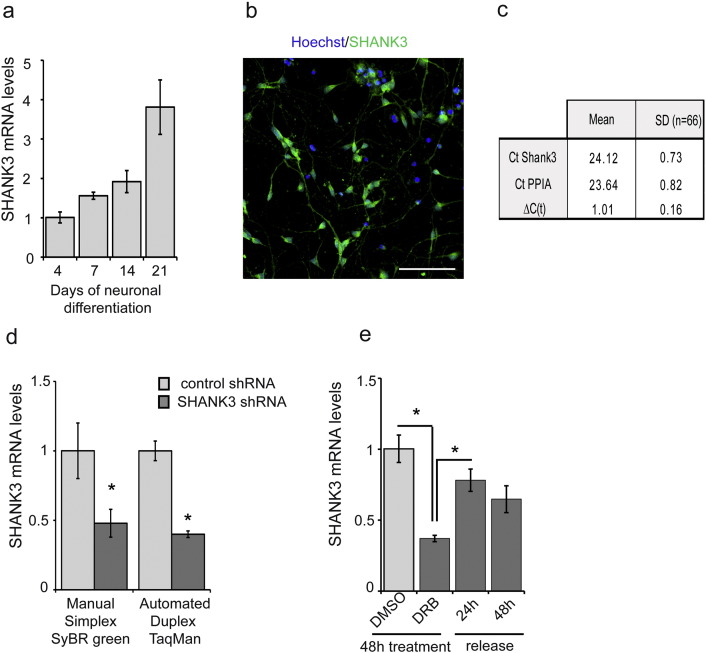
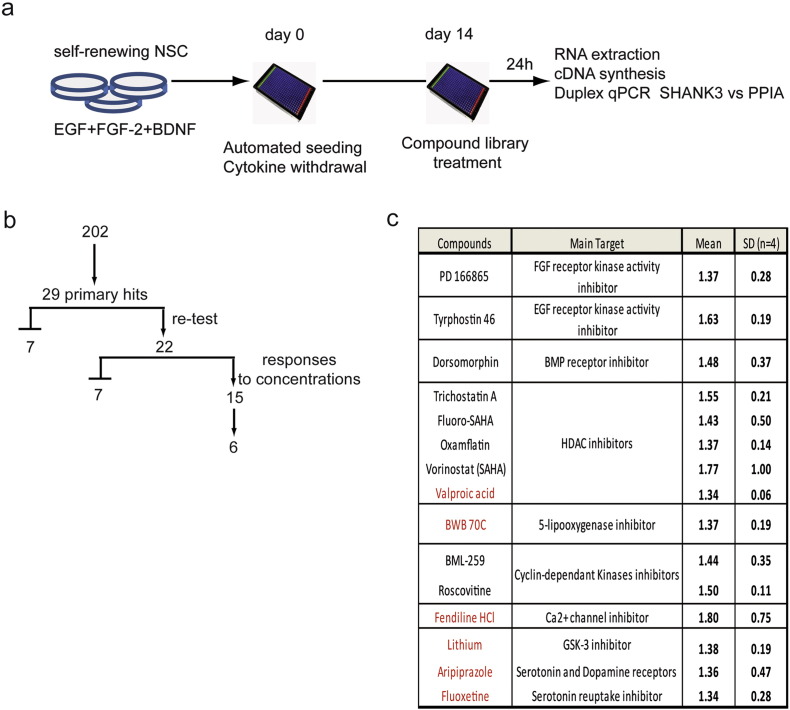
To start cortical neuron differentiation, NSC were dissociated and plated in N2B27 at a density of 6000 cells/wells in poly-l-ornithin/laminin pre-treated 384 well plates using the Bravo™ automated liquid handling platform equipped with a 384 pipette head (Agilent Technologies Inc., Santa-Clara, CA, USA). Half of the medium was changed twice a week. In those conditions, neuronal differentiation is achieved after 11 to 14 days. Kinetic of SHANK3 mRNA expression was established to determine the best time point to perform the screening (Fig. S1) and day 14 was chosen since it offered the best dynamic range to quantify increase in SHANK3 mRNA expression. At day 14, compounds to be tested were added using the same platform in four replicates. The development and validation of the automated high throughput PCR assay by itself was performed following published standards for qPCR based HTS (Bittker, 2012). mRNAs were extracted and directly converted in cDNA in the assay plate using the Fastlane technology (FastLane Cell Multiplex Kit, Qiagen, Courtaboeuf, France) with minor modifications. A duplex qPCR strategy was used, consisting in amplifying and quantifying both SHANK3 and the house keeping gene PeptidylProlyl Isomerase A (PPIA) in the same well (QuantiFast Probe Assay DP Kit). The robustness of this method was evaluated by performing it in 66 technical replicates. Ct values were consistent between technical replicates with minimal inter-well variations (Fig. S1). In order to evaluate the sensitivity of the duplex TaqMan PCR to SHANK3 mRNA level modifications, SHANK3 specific shRNAs were used to artificially decrease the level of transcripts in PSC-derived neurons. Sister neuronal cultures were nucleofected (Amaxa, Lonza, Amboise, France) with non-targeting shRNAs as controls. The performance of the simplex method was compared with the automated duplex Taqman qPCR strategy and both strategies found equally efficient, indicating than automation and multiplexing had not compromised the sensitivity and specificity of the assay (Fig. S1). Finally, the dynamics of SHANK3 mRNA synthesis was analyzed in order to estimate a relevant duration of treatment with the tested compounds. Neurons were treated with 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole (DRB) during 48 h to block mRNA production (Fig. S1 Sehgal et al., 1976). Treatment with 100 μM of DRB decreased SHANK3 mRNA content by 63%. After 24 h of release from DRB, SHANK3 mRNA contents were partially reconstituted and reached 80% of the non-treated cells. Neurons were, therefore, treated over 24 h for drug screening. The final screening workflow is summarized in Fig. 1a. Fold change variations induced by compound treatments were calculated using the 2ΔCt method with DMSO-treated cells as controls and PPIA as the reference gene.
Fig. S1.
SHANK-3 screening assay development
a) Quantification of SHANK-3 mRNA in hESC-derived differentiating neurons using manual mRNA extraction and SYBR green detection techniques. b) Expression of SHANK-3 protein in neurons at day 14. Scale bar = 200 μm c) Mean Ct values and standard deviations of SHANK-3 and PPIA detection using Duplex Taqman method evaluated in 66 technical replicates of DMSO treated neurons. ΔCt corresponds to Ct PPIA-Ct SHANK-3. d) Comparison of the sensitivity of the manual RNA extraction following by simplex SYBR green detection method with the automated RNA extraction combined with TaqMan duplex detection method using neurons transfected with control or SHANK-3 specific shRNAs. Results represent mean ± SD of 4 independent biological replicates. * p < 0.001 t-test. e) Determination of SHANK-3 mRNA turn over using the mRNA synthesis inhibitor 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB). Results represent mean ± SD of 3 independent biological replicates. * p < 0.001 t-test.
Fig. 1.
SHANK3 screening results.
a) Screening work flow. b) Screening success rate. Among 202 molecules, 29 primary hits (increase of at least 30% of expression of the DMSO-treated wells which correspond to a statistical deviation of more than 2 σ) were identified. Independent re-testing confirmed 22 compounds. Dose response experiments revealed a concentration-dependent mode of action for 15 compounds. Among this list of 15 compounds, 6 were chosen for further investigation. c) Table summarizing the name of the 15 compounds demonstrating a dose-dependent effect on up-regulating SHANK3 mRNA, their known biological target or medical application and their performance in the primary screening. Results are presented as the mean fold-change values compared to DMSO treated cells and standard deviations (SD) of the 4 biological replicates. The 6 compounds chosen for further investigation are in red.
2.5. Automated Image-based High Content Screening
For high-content screening assays, cells were fixed in 4% paraformaldehyde for 15 min at 4 °C. Primary antibodies (Table S2) diluted in blocking buffer were applied overnight at 4 °C. Hoechst 33,258 (2 μg/ml, Sigma) and secondary antibodies conjugated to Alexa fluorophores (Molecular Probes, Eugene, Oregon, United States) were diluted at 1:1000 in blocking buffer and applied for 2 h at room temperature. To quantify synapses, image acquisition and analyses were performed using the ImageXpress® Micro XLS system (Molecular Devices, Sunnyvale, CA, USA). A presynaptic marker and a postsynaptic marker were used and multiplexed with the neuronal markers HuC/D and beta III Tubulin (anti-Tuj-1 antibodies) to identify neurons and create a “neurite” mask. An algorithm was then generated using MetaXpress™ software application modules in order to identify pre- and post-synaptic spots specifically located on neurites, then to count the number of spot co-localization (considered as synapses). Results were expressed as the density of spots for 100 μm of neurite. A minimum of 10 sites per well (approximately 2000 neurons) was analyzed. For neuritic network parameter analysis, image acquisition was performed using Cellomics Arrayscan system (Thermofischer Scientific, Waltham, MA, USA). Beta-III tubulin staining was used to track neurite using “Neuronal profiling” bioapplication as described in (Boissart et al., 2013).
2.6. Spontaneous Calcium Oscillation Recording
After 35 to 42 days of differentiation, neurons were treated during 120 h with compounds of interest in 384 well-plates. Cells were incubated for 10 min at 37 °C with the Fluo-4 probe diluted at 1 mM in a loading buffer (HBSS 1X, Hepes Buffer 20 mM, pH 7.4). After 3 washes with Mg2 +-free Buffer (HBSS w/o CaCl2, w/o MgCl2, Hepes Buffer 20 mM, CaCl2 2 mM, pH = 7.4), neurons were recorded in the Mg2 +-free Buffer. The fluorescence reading was performed with the ImageXpress® Micro XLS System at 37 °C. Neurons were recorded every second during 3 min with an exposure time of 350 ms. Total fluorescence curves were extrapolated using MetaXpress software.
2.7. Transcriptome Analysis
For transcriptome analysis of gene expression perturbations induced by lithium and VPA, human neurons differentiated from the SA001 line were treated during 24 h with compounds and RNA extracted using RNAeasy extraction kit following manufacturer's instructions (Qiagen). Fifty ng of total RNA was reverse transcribed using the Ion AmpliSeq Transcriptome Human Gene Expression kit (Revision A.0) following the protocol of the manufacturer (Thermo Fisher Scientific). The cDNA libraries were amplified and barcoded using Ion AmpliSeq Transcriptome Human Gene Expression core panel and Ion Xpress Barcode Adapter (Thermo Fisher Scientific). The amplicons were quantified using Agilent High Sensitivity DNA kit before the samples were pooled in sets of eight. Emulsion PCR and Enrichment and loading was performed on the Ion Chef Instrument using the Ion PI IC 200 Kit (Thermo Fisher Scientific). Samples were loaded on an Ion PI v3 Chip and sequenced on the Ion Proton System using Ion PI IC Hi-Q 200 Kit chemistry (200 bp read length Thermo Fisher Scientific). The output files (FASTQ files) were imported into the RNA-seq pipeline of Partek Flow software (v 4.0, Partek Inc., St Louis, MO, USA) using hg19 as a reference genome. To determine genes that are differentially expressed between groups mapped reads were quantified using Partek E/M algorithm (the resulting counts represent the gene expression levels for over 20,800 different genes present in the AmpliSeq Human Gene Expression panel) and differentially expressed gene were identified using Partek Gene Specific Analysis (GSA) algorithm. Biological interpretations of the list of differentially expressed genes were performed using Partek Flow v4.0 and the pathway analysis plateform EnrichR (Chen et al., 2013).
2.8. Statistical Analyses
Data were subjected to ANOVA following by a t-test to evaluate statistical differences between cells treated with tested compounds and DMSO-treated controls. The number of biological replicates, corresponding to independent differentiations or neuronal cultures, is indicated in figure legends.
3. Results
3.1. Screening of Compounds Regulating SHANK3 mRNA Expression in Human Neurons Differentiated from hESC
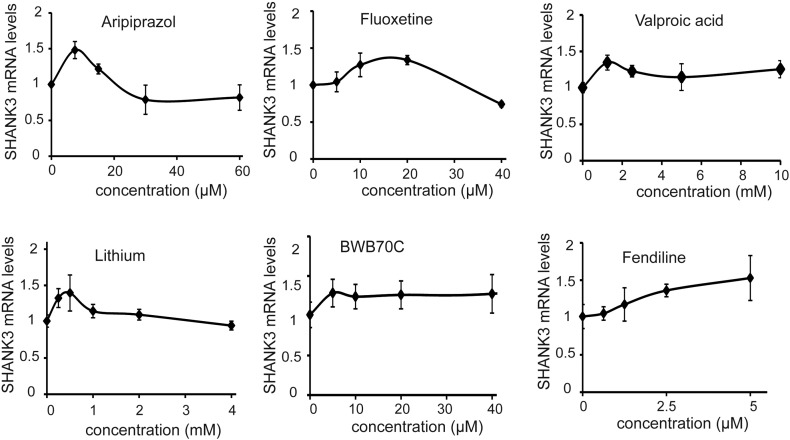
A collection of 202 compounds was chosen for the primary screening of SHANK3 regulators. This collection included a set of FDA-approved therapeutic compounds widely used for a variety of psychiatric and neurological diseases including epilepsy, bipolar disorders, major depressive disorders or schizophrenia. It also included epigenetic regulators, kinase inhibitors and compounds identified previously as modulators of neurogenesis (Boissart et al., 2013) (Table S1). Reprogramming of somatic cells into iPSC being potentially responsible for induction of epigenetic and imprinting alterations that could bias the study of gene regulation (Pick et al., 2009, Urbach et al., 2010, Lister et al., 2011), the primary drug screening was conducted using neurons differentiated from the control hESC line SA001. The 202 compounds were screened in 4 biological replicates at one concentration determined for each compound based on data from the literature or from our own previous findings (Boissart et al., 2013, Georges et al., 2015). This primary drug screening identified 29 primary hits that increased SHANK3 mRNA content by at least 30% as compared to DMSO-treated cells and with a deviation from the DMSO mean of more than 2 σ (Fig. 1b and Table S1). Re-testing of these 29 hits in an independent experiment confirmed 22 compounds (Fig. 1b). Dose-response experiments were then conducted and revealed a concentration-dependent mode of action for 15 of them (Fig. 1b and c). Among this list of 15 compounds, 6 were chosen to be further investigated: 5 FDA-approved compounds (fendiline dihydrochloride, lithium, valproic acid, aripiprazol and fluoxetine) and the 5-lipoxygenase inhibitor BWB-70C since it regulated a pathway that can be targeted by the FDA-approved compound Zileuton. The optimal concentrations for each pharmacological compound were determined from the mRNA dose-response curves, and defined as follows: lithium 0.5 mM, valproic acid (VPA) 2 mM, aripiprazol 7.5 μM, fluoxetine 10 μM, fendiline 5 μM, BWB70C 40 μM (Fig. S2).
Fig. S2.
Concentration – response curves of the 6 chosen compounds.
Results are expressed as Fold-change compared to DMSO-treated cells and presented as mean ± SD of 3 independent replicates.
3.2. Modification of SHANK3 Synaptic Content and Neuronal Activity by Hit Compounds
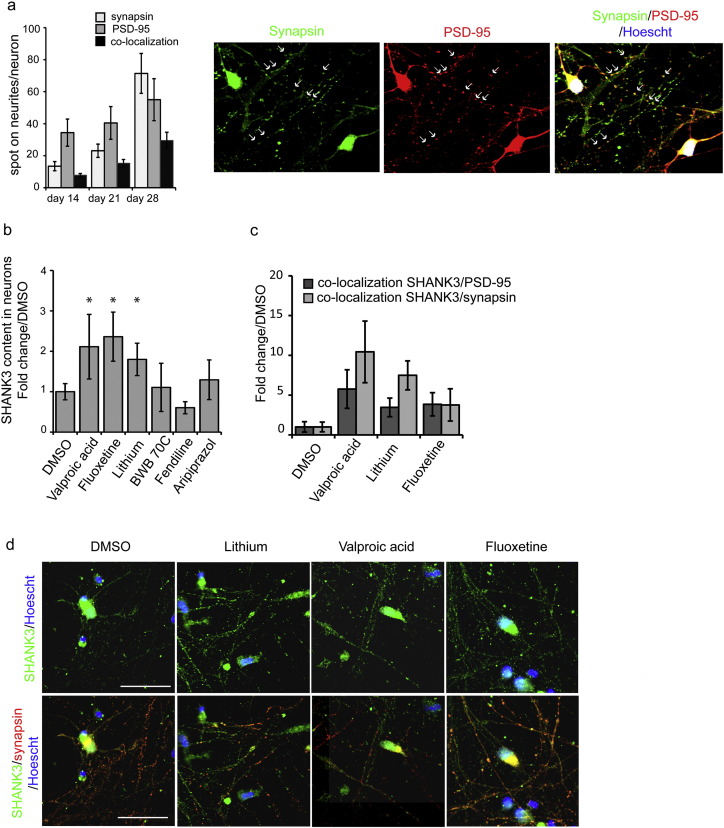
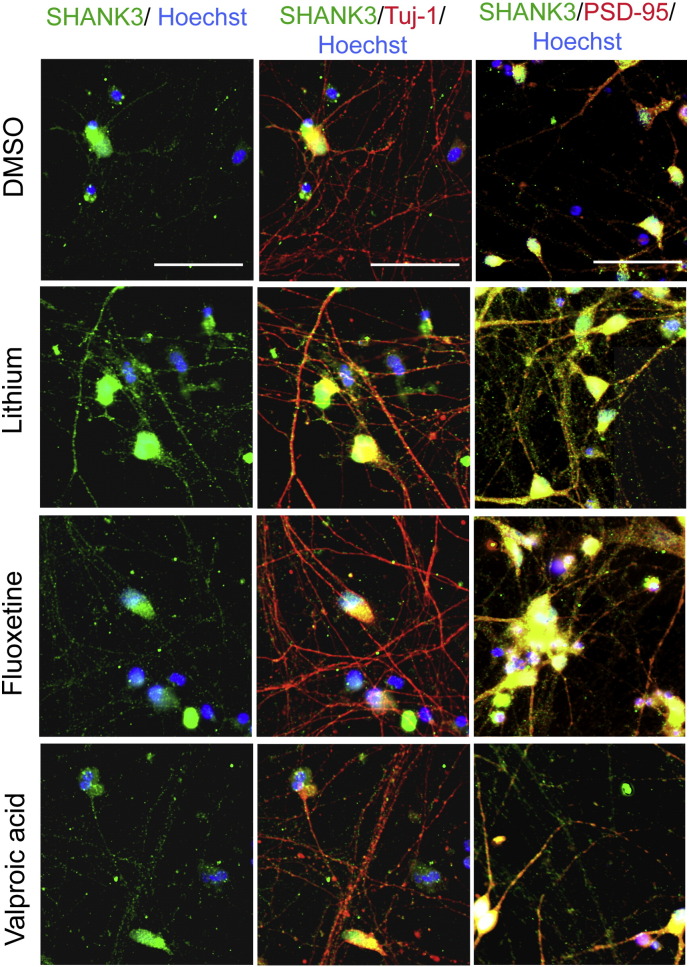
The functional potential of the six drugs was first evaluated in neurons derived from the SA001 hESC line. SHANK3 protein levels and its recruitment to the glutamatergic synapses, as well as increased glutamatergic activity of neuronal network were used as indicators of compound efficacy. The time course of synaptogenesis was analyzed by quantifying the number of spots of the pre-synaptic protein Synapsin and of the post-synaptic density component PSD-95 present on neurites, a co-localization suggesting a glutamatergic synapse (Fig. 2a). In untreated hESC-derived neuronal cultures, expression of Synapsin and PSD-95 in the neuritic compartment progressively increased from day 14 to 28 and synapses were clearly present at day 28. The effects of the 6 compounds on SHANK3 synaptic content was therefore analyzed at that later stage. In order to optimally quantify SHANK3 protein production following treatment, cells were exposed to compounds for at least 120 h.
Fig. 2.
Effect of hit compounds on SHANK3 protein expression and synaptic location.
a) Time course analysis of synapse formation in hESC-derived developing neurons. Results are expressed as mean ± SD of 3 independent neuronal cultures (biological replicates, left panel). Representative images of the pre-synaptic marker Synapsin (green) and the post-synaptic marker PSD-95 (red) are shown on the right panels. Arrow indicates example of co-localization of PSD-95 and Synapsin signaling structural synapses. b) Quantification of SHANK3 protein expression on dendrites after treatment with hit compounds. Results are expressed as mean ± SD of 6–8 independent neuronal culture/ treatment (biological replicates) and as fold change compared to DMSO-treated cells. * p < 0.01 t-test c) Quantification of SHANK3 protein co-localization with synapsin and PSD-95 after compound treatment. Results are expressed as fold change of DMSO treated cells (mean ± SD, 6–8 biological replicates par treatment). d) Representative image of SHANK3 (green) co-localization with synapsin (red). Scale bar = 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Only 3 out of the 6 compounds increased SHANK3 at the protein level in neurons at the tested concentrations, namely lithium, VPA, and fluoxetine (Fig. 2b). The same 3 compounds also significantly increased the number of neuritic sites in which SHANK3 co-localized with PSD-95 (Fig. 2c and d; Fig. S3), suggesting an efficient recruitment at the post-synaptic density. Treatment with lithium and VPA, but not fluoxetine, increased the co-localization of SHANK3 with the presynaptic marker Synapsin, taken as the indication of SHANK3 containing synapses (Fig. 2c and d).
Fig. S3.
Expression of SHANK-3 in hESC derived neurons.
Representative images of SHANK-3 co-localisation with the neuritic marker Tuj-1 and the post-synaptic protein PSD-95. Scale bar = 50 μm.
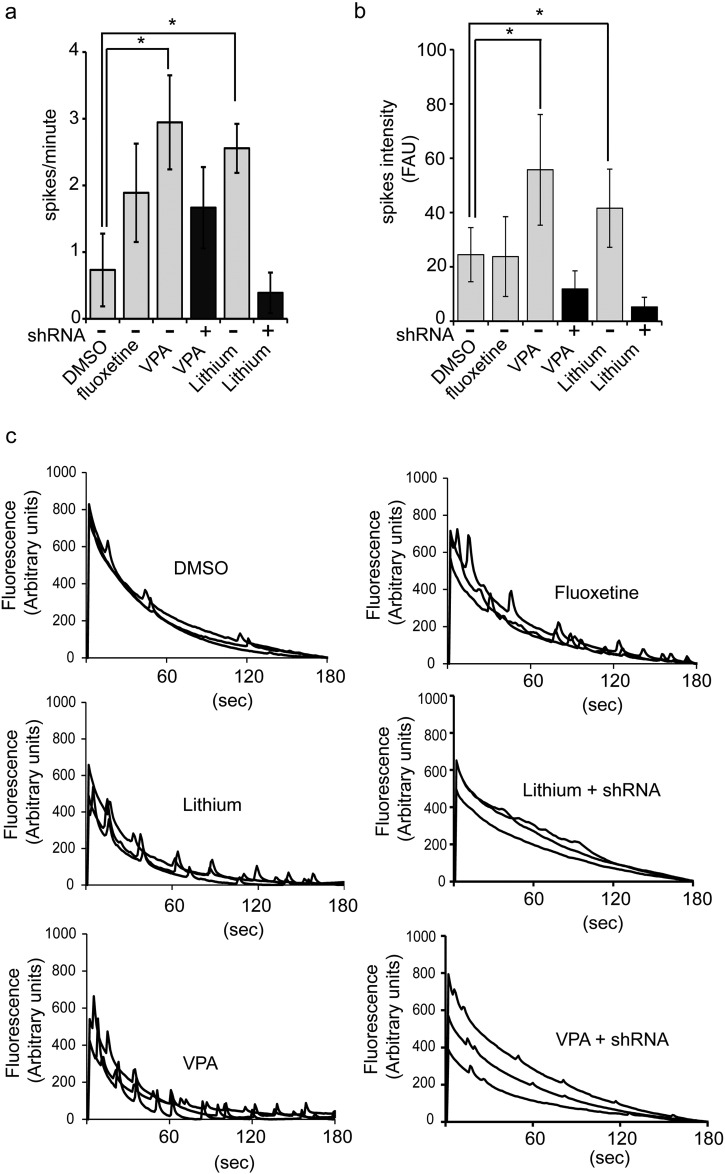
The relationship between the increased number of SHANK3-containing synapses and functional changes in the neuronal network connectivity was then investigated. At day 37, neurons were treated with lithium, VPA or fluoxetine during 5 days, then network spontaneous calcium oscillations (SCO), reflecting the number of mature synapses (Dravid and Murray, 2004, Cao et al., 2015), were measured using Fluo-4 calcium probe (Fig. 3). Lithium and VPA, but not fluoxetine, increased both SCOs frequency and intensity in treated cortical neurons in a statistically significant manner indicating increased network connectivity. This effect was strongly decreased (VPA) or totally lost (lithium) in neurons in which shRNAs targeting SHANK3 were expressed, confirming that the effects of the drugs were dependent on SHANK3 (Fig. 3).
Fig. 3.
Functional analysis of neuronal network activity following compounds treatment.
a–b) Quantification of spontaneous calcium oscillations (SCO) frequency (spikes/min, a) and intensity (FAU: Fluorescence Arbitrary Units, b) in neurons treated with DMSO, fluoxetine, lithium and valproic acid (VPA). SCO were recorded over a period of 180 s, results represent mean ± SD of 5–6 biological replicates/treatment. *p < 0.01 t-test compared to DMSO treated cells. shRNA: neurons stably expressing a cDNA coding for a SHANK3 shRNA c) Representative example of recordings.
3.3. mRNA Profiling Following VPA and Lithium Treatments
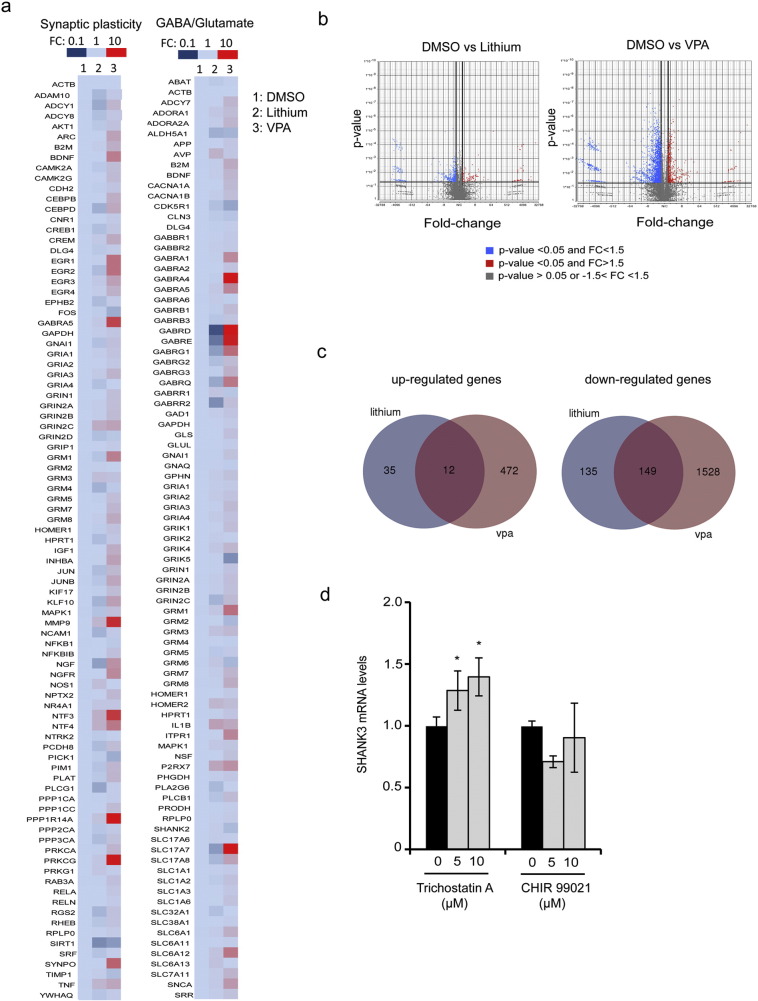
In order to document more generally the effect of lithium and VPA on synaptic mRNAs, expression profile of 150 neuronal and synaptic genes was established after treatment of SA001-derived neurons by lithium or VPA (Fig. 4a and Table S3). Lithium exerted a discrete effect with only 16 genes up-regulated in the same range as SHANK3, i.e. with a fold change compared to DMSO of more than 1.4, none of them being up-regulated more than 3 fold. Among these 16 genes, 6 were direct or indirect partners of SHANK3 at the synapse, namely, the PSD protein HOMER2, the metabotropic glutamate receptors GRM1, GRM3 and GRM6, the NMDA receptor sub-unit NR4A1 and the metalloprotease MMP-9. The 10 additional genes were neurotrophins (NTF3 and NTF4), GABA transporters (SLC6A12 and SLC6A13), one purinergic receptor (P2RX7), interleukin IL1B, arginine vasopressin AVP, NO synthase NOS-1 and two protein kinases (PRKG1 and PRKCG). In contrast, VPA profoundly modified synaptic mRNAs content with more than 60% of the genes being up-regulated above 1.5 fold as compared to DMSO, and up to 34 fold changes. This discrete effect of lithium on gene expression was further confirmed by global transcriptomic analysis. Whole genome RNA sequencing revealed that only 331 genes were modified by lithium treatment in SA001 neurons (Fold change >+1.5 or − 1.5 compared to DMSO treated neurons, p < 0.05). In contrast, VPA treatment induced the dysregulation of 2161 genes in the same range (Fig. 4b, c and Table S4).
Fig. 4.
Molecular targets of VPA and Lithium in human neurons.
a) Heat map of gene expression profiles of 150 genes involved in synaptic plasticity or GABA/glutamate pathways in DMSO (column 1), lithium (column 2) or valproic acid (VPA, column 3) treated neurons. FC: Fold change of mRNA expression when compared to DMSO-treated cells. b) Volcano plots showing the distribution of differentially expressed genes in neuronal cultures treated with Lithium or VPA compared to DMSO (p value GSA test < 0.05). Results represent mean of 3 independent neuronal cultures per treatment. c) Venn's Diagram illustrating genes independently or commonly targeted by lithium and VPA treatments. d) Effect of inhibitors of GSK-3 (CHIR 99021) and HDAC (Trichostatin A) dependent pathways on SHANK3 mRNA expression. SHANK3 mRNA levels were quantified 24 h after treatment with 5 or 10 μM of the indicated inhibitor. PPIA was used as a reference gene. Results are expressed as fold change compared to DMSO treatment (0 μM) and represent mean ± SD of 4 independent neuronal cultures per treatment. * p < 0.05.
3.4. Up-regulation of SHANK3 mRNA is dependent of HDAC inhibition but independent on GSK-3 inhibition
Candidate signaling pathways potentially linking VPA and lithium to SHANK3 up-regulation were further investigated. The way each one of these pharmacological agent exerts its therapeutic activity on neuronal cells is not fully understood since both are suggested to inhibit a large variety of enzyme-dependent cascades (Gould et al., 2004). We therefor decided to investigate commonly targeted signaling pathways. Lithium and VPA are considered as two mood stabilizers sharing at least two important common cellular targets: the Glycogen Synthase Kinases of type 3 (GSK-3) and class I Histone Deacetylases (HDAC) (Wu et al., 2013, Lee et al., 2015, Chen et al., 1999). Involvement of these two pathways in SHANK3 mRNA regulation was investigated by treating SA001-derived neurons using Trichostatin A, a potent and selective HDAC inhibitor, or CHIR99021 a selective GSK-3 inhibitor (Fig. 4d). After 24 h of treatment, Trichostatin A significantly increased SHANK3 mRNA. In contrast, CHIR99021 was not able to mimic the effect of lithium and VPA on SHANK3 mRNA. In parallel, RNA sequencing data indicated that lithium and VPA commonly up-regulated 12 genes while down-regulating 149 (Fig. 4c, Table S4). This list of gene was further investigated using the pathway enrichment analysis tool PANTHER (Protein ANalysis THrough Evolutionary Relationships) via EnrichR. Commonly regulated genes did not cluster into signaling pathways linked to GSK-3 or any metabolic enzymes directly targeted by lithium or VPA. In contrast, among the 149 down-regulated genes, 5 histone isoforms were identified (HIST1H1B, HIST1H1C, HIST1H2B, HIST1H4A, HIST1H4D, Table S4). Together, this suggested that lithium and VPA-mediated increase of SHANK3 mRNA levels could be triggered via an epigenetic mechanism involving histone regulation.
3.5. Lithium and VPA Action on Neurons Differentiated from iPSC of Patients with SHANK3 Mutations
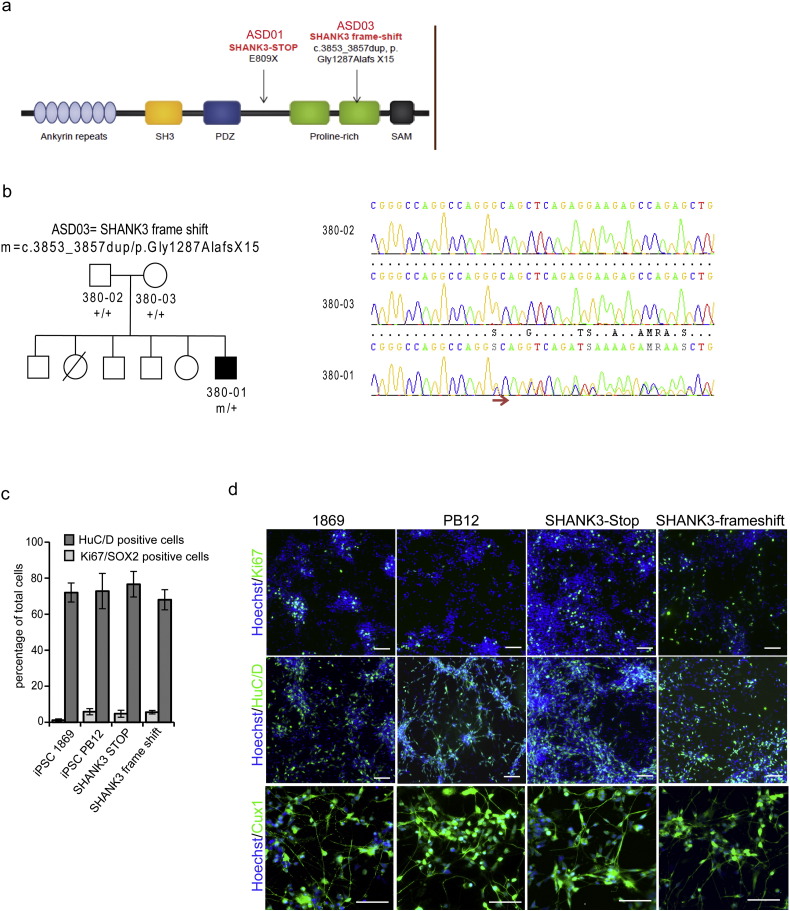
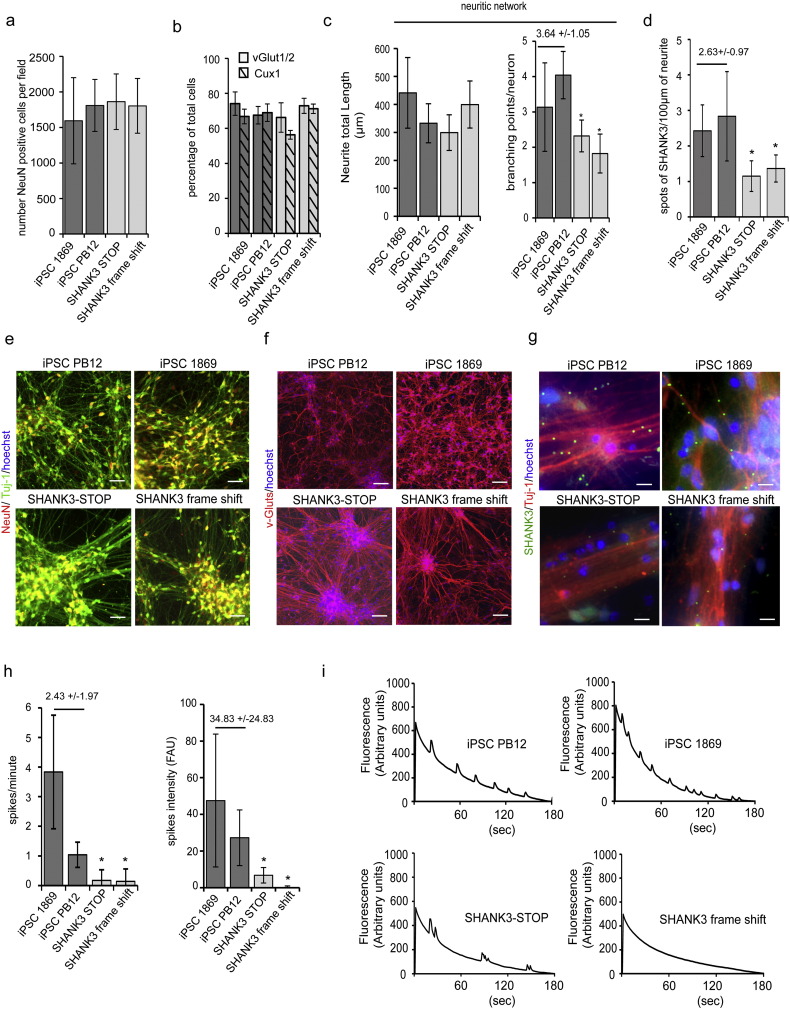
To evaluate compounds efficacy on SHANK3 haplotypes associated phenotypes, iPSC lines were derived from two children with autism and severe intellectual disabilities who carried two different deleterious mutations in one allele of SHANK3 (SHANK3-STOP and SHANK3 frame-shift, Fig. S4 and Leblond et al., 2014). The two mutations were predicted to lead to loss of function of one of the two alleles of SHANK3, leaving one functional allele. Neurons were first differentiated and compared to neuronal cultures differentiated from two control lines PB12 and 1869 (Fig. 5 and Fig. S4). As described previously, SHANK3 haplo-insufficiency did not significantly influence the rate of neuronal differentiation as determined by quantifying the fraction of the cells expressing the neuronal specific proteins HuC/D (Fig. S4) or NeuN (Fig. 5a and e) and Tuj-1 (Fig. 5e). After 28 days of differentiation, all the culture contained more than 70% of post-mitotic neurons independently of the presence or absence of SHANK3 haplo-insufficiency. In parallel, the fraction of dividing neuronal precursors, characterized by the co-expression of Sox2 and Ki67 were lower than 10% (Fig. S4). SHANK3 haplo-insufficiency did not modify the type of neurons produced that were mostly glutamatergic neurons, expressing vGlut-1/2 (Fig. 5b and f), of the superficial cortical layers (expressing the layer specific marker Cux1, Fig. 5b and Fig. S4, Boissart et al., 2013, Yi et al., 2016). Neuronal phenotypes previously associated with SHANK3 loss of function were then deeply analyzed to determine whether they were robustly recapitulated in neurons differentiated from iPSC of patients with disruptive point mutation in SHANK3 gene. Neuritic network morphology was analyzed by automatically tracking neurite using beta-III tubulin staining (anti-Tuj-1 antibodies, Fig. 5c and e). Analysis of two different neuritic network parameters, i.e. the size of the neuritic network associated with a single neuron (neurite length by neuron) and the way this neuritic network develop complexity (branching point number per neuron) indicated that SHANK3 haplo-insufficiency impacted significantly neurite branching (Fig. 5c and e). Quantification of the density of SHANK3 spots on neurites confirmed that SHANK3 haploinsufficiency was associated with lower SHANK3 expression at synaptic sites since the density was significantly lower in the neuronal cultures differentiated from the two SHANK3 mutated iPSC lines (Fig. 5d, g). Together, this led to deficient neuronal connectivity as revealed by decreased intensities and frequencies of SCOs recorded in the two patient-derived neuronal cultures after 42 days of differentiation (Fig. 5h, i).
Fig. S4.
Additional characterization of iPSC-derived neurons and response to lithium and VPA treatments.
a) Schematic representation of SHANK3 protein domains and description of the two SHANK3 mutations identified in the ASD children from whom iPSC were derived. b) Pedigrees of the family of patient ASD03 (SHANK3 frameshift). The chromatograms were obtained after Sanger sequencing. The arrow indicates the frame-shift. c) Quantification of neurogenesis, in control (1869 and PB12) and SHANK3 mutated (SHANK3 STOP and SHANK3 frame shift) lines. Post-mitotic neurons were quantified using the neuronal specific nucleo-protein HuC/D whereas SOX2/ki67 double positive cells represent cycling neuronal precursors. Results represent mean ± SD of at least 2 independent differentiations. d) Representative expression of HuC/D, Ki67 and Cux1 in 28 days neuronal cultures from the two control lines PB12 and 1869 and from the two SHANK3 mutated lines. Scale bars = 50 μm.
Fig. 5.
Neurons differentiated from patients with SHANK3 mutations recapitulate cellular phenotypes associated to SHANK3 haploinsufficiency.
a) Quantification of the number of neurons (NeuN/Tuj-1 double positive cells) per field of 330 μm2, in control (1869 and PB12) and SHANK3 mutated (SHANK3 STOP and SHANK3 frame shift) lines. Results represent mean ± SD of at least 6 independent differentiations, with a minimum 10 fields per culture. b) Quantification of neuronal sub-type present in 28 days neuronal culture obtained from controls or SHANK3 cells. vGlut1/2 expression indicated glutamatergic neurons and Cux-1 expression marked cortical glutamatergic neurons of the superficial layers. Results are mean ± SD of 3 independent differentiations. c) Quantification of neuritic network parameters in control (iPSC 1869 and iPSC PB12) and SHANK3 mutated (SHANK3-STOP and SHANK3 frame shift) 28 days-old neuronal cultures. Neurite total length represents the size in μm of the neuritic network associated with one neuron while the number of branching points quantifies more specifically neurite network complexity. Results represent mean ± SD of at least 6 independent neuronal cultures per line. d) Quantification of SHANK3 synaptic content in neurons derived from control (1869 and PB12) or SHANK3 mutated lines. Data are reported as the density of spots per 100 μm of neurite. Results represent mean ± SD of 4 independent neuronal cultures per line. e) Representative images of the neuronal density (NeuN positive cells) and of the neuritic network revealed using beta-III tubulin staining (Tuj-1 antibodies) in control and SHANK3 neuronal cultures. Scale bar = 50 μm. f) Representative images of the v-Gluts (mixed antibodies recognizing v-Glut1 and vGlut-2) staining in control and SHANK3 neuronal cultures. Scale bar = 100 μm. g) Representative images of SHANK3 spots on neurite of control and SHANK3 neurons. Scale bar = 20 μm. h) Quantification of spontaneous calcium oscillation frequency (spike/min) and intensity (F.A.U: Fluorescence Arbitrary Units), reflecting neuronal network activity, in control (PB12 and 1869) and in SHANK3 mutated (SHANK3-STOP and SHANK3 frame-shift) in 42 days-old neuronal culture. Results represent mean ± SD of recording of at least 6 neuronal cultures per lines. In panel c, d and h, values above the two dark grey bars represent the mean ± SD of the two control lines. *: p < 0.05, control lines vs the indicated SHANK3 mutated line. i) Representative recording of calcium oscillations in control and SHANK3 neuronal cultures.
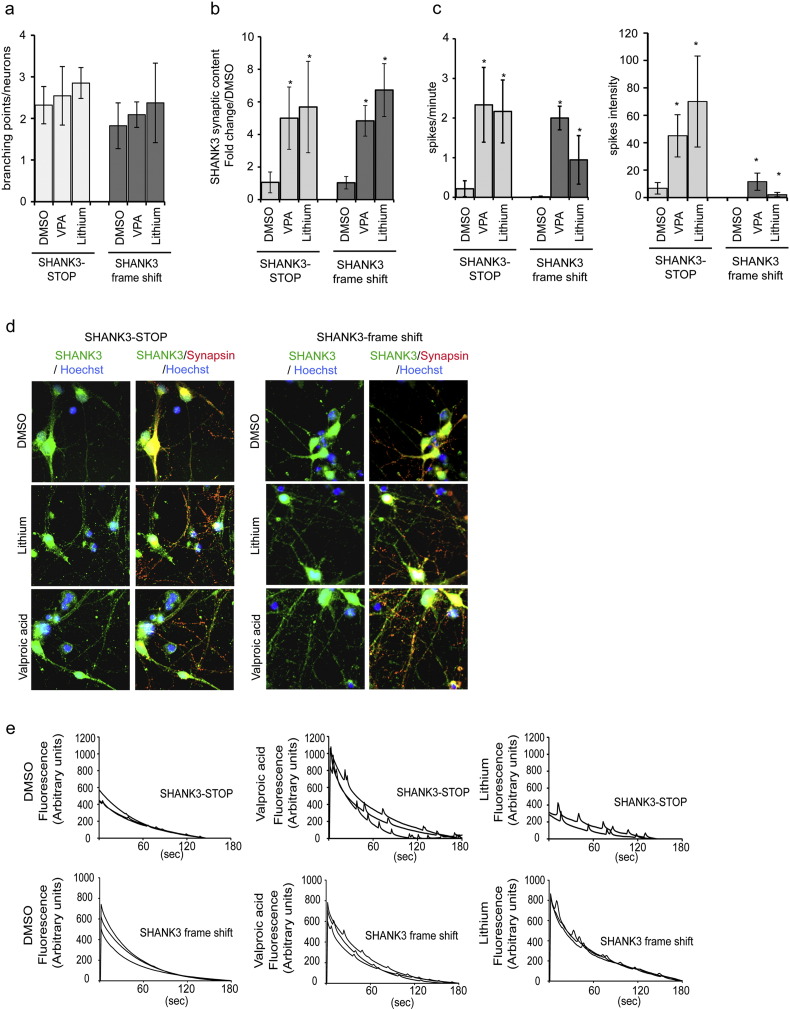
The recapitulation of SHANK3 deficiency associated phenotypes in neurons of patients with disruptive point mutations allowed us to use them as a biological support to test the efficiency of VPA and lithium. Neurons with SHANK3 mutations were treated with VPA or lithium during 120 h. The two drugs had only a slight effect on neurite branching (Fig. 6a) but significantly increased the number of SHANK3-containing synapses (Fig. 6b, d) as well as SCOs frequency and intensity in the two SHANK3 genetic backgrounds (Fig. 6c, e). This suggested that these two compounds may be of beneficial for patients from whom the iPSC were derived.
Fig. 6.
VPA and Lithium treatment efficiency in neurons with SHANK3 mutations.
a) Quantification of the number of neurite branching points of neurons derived from SHANK3 mutated lines after lithium and VPA treatments. Results represent mean ± SD of 3 independent differentiation/treatment. b) Quantification of SHANK3 synaptic content in neurons derived from iPSC individuals bearing the two different SHANK3 mutations (SHANK3-STOP and SHANK3 frame shift) after 120 h treatment with valproic acid (VPA) or lithium. Results are presented as fold change variations compared to DMSO-treated cells and represented as mean ± SD of at least 6 independent biological replicates/treatment. c) Quantification of spontaneous calcium oscillation frequency and intensity in SHANK3 mutated lines (SHANK3-STOP and SHANK3 frame-shift) with or without treatment with VPA or lithium. Data represent mean ± SD of 4 independent neuronal cultures per treatment. * p < 0.01 Lithium or VPA vs DMSO. d) Representative images of SHANK3 and Synapsin expression in DMSO, VPA and Lithium treated neurons. e) Representative recording of calcium oscillations.
3.6. Clinical Assessment of Lithium Treatment on ASD Severity in a SHANK3-STOP Patient
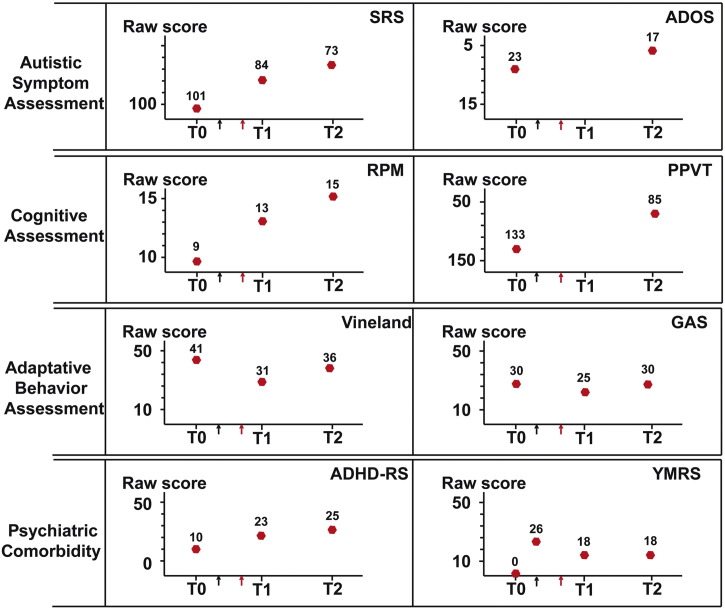
Patient AUN-003, from whom the iPSC line SHANK3-STOP was derived (de novo truncating mutation P.Glu809X) was enrolled in 2012, aged 12 years old, in an observational clinical study (Fig. 7, T0). She had a severe ASD measured with the Social Responsiveness Scale (SRS, raw score = 101, t-score = 89), the Autism Diagnosis Observational Scale (ADOS, social affect domain raw score = 23) and the Autism Diagnosis Interview-Revised version (data not shown). She displayed a mild intellectual disability with contrasting results on the Peabody Picture Vocabulary Test (PPVT) (t-score: 101) and the Raven's progressive matrices (RPM) (raw score = 9, t-score: < 65). However, her adaptive behavior was below her cognitive ability based on the Vineland's module 2 (communication domain raw score = 41) or the Global Assessment Scale (GAS, raw score = 30). At T0, by screening her psychiatric comorbidity with the Kiddie SADS, a semi-structured interview, patient AUN-003 had no lifetime history of Attention Deficit Hyperactive Disorder (ADHD) or of bipolar affective disorders (including manic symptoms). Seven months later (Fig. 7, black arrow), she developed symptoms of mood deregulation. During several months, she displayed major depressive symptoms followed by a hypomanic episode with mixed features. The maximum intensity of manic symptoms was retrospectively assessed with the Young-Mania rating Scale (Y-MRS, raw score = 26). Five months later, (Fig. 7, red arrow), facing inability to control the symptoms, lithium was introduced as an indication for bipolarity. Considering the in vitro results obtained on her iPSC derived neurons, ASD scores were carefully assessed. After 8 months of treatment (T1), she displayed a clear improvement on manic symptoms (Y-MRS raw score = 18). In addition, a modest but significant positive effect was identified on ASD severity, as evaluated by the SRS (raw score = 84, t-score = 80), as well as on her cognitive performance measured with the RPM (raw score = 13, t-score < 70). However, the adaptive behavior was not improved whatever the scale considered (GAS = 25, Vineland communication domain score = 31). After one year (T2) adaptive behavior started to improve as compared to T1, though still close to the performance at T0 (GAS = 30, Vineland score = 35), i.e. before regression. In contrast, the caregivers reported additional reduction of autistic symptoms on the SRS (raw score = 73, t-score = 75), which was further confirmed by results on the ADOS (social affect domain raw score = 17). These two tests revealed that ASD symptoms were lower than before regression. In contrast, there has been a degradation of her ability to control her impulsivity between T1 and T2, associated to distractibility. Caregivers mentioned that these symptoms differed from those observed during the manic episodes. Despite their late onset, symptoms tended to evoke clinical features of ADHD (ADHD-RS, global raw score = 25). Table S5 reports in details the scores obtained for each tests and each sub-categories.
Fig. 7.
Clinical assessment of patient AUN-003 (SHANK3-STOP).
T0: Patient AUN-003 aged 12 years old, was enrolled in the study (2012). Black arrow: she developed symptoms of mood deregulation and regression. Red Arrow: start of lithium treatment (0.6 mL–1000 mg/d). T1: 8 months follow-up, T2: one year follow-up. SRS: Social Responsiveness Scale, ADOS: Autism Diagnosis Observational Scale, RPM: Raven's progressive matrices, PPVT: Peabody Picture Vocabulary Test, GAS: Global Assessment Scale, ADHD-RS: Attention deficit associated to hyperactivity disorder rating Scale, Y-MRS: Young-Mania rating Scale. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
SHANK3 haplo-insufficiency induced by heterozygous loss of function mutations represents a clear model of abnormal gene dosage leading to ASD (Durand et al., 2007, Yi et al., 2016) that can be reversed by genetically re-introducing SHANK3 normal levels of expression, including during adulthood, in mice models (Shcheglovitov et al., 2013). (Mei et al., 2016). We therefore proposed that successful therapeutic compounds for SHANK3 haplo-insufficiency dependent form of ASD could be identified based on their potential to up-regulate this gene in human neurons. We first selected potential hit compounds using control hESC derived neurons, to avoid epigenetic bias potentially induced by iPSC reprogramming (Urbach et al., 2010, Lister et al., 2011). Patient-derived iPSC cellular models were then used to challenge their efficacy at corrected SHANK3-associated neuronal phenotypes considered as relevant for ASD symptoms (Bozdagi et al., 2010, Yi et al., 2016). Interestingly, among the compounds regulating SHANK3 identified in the present study were the two mood stabilizers VPA and lithium that share common cellular targets. Additionally, we reported promising results of lithium pharmacotherapy to reduce ASD core symptoms in one of the patient from whom iPSC were produced. Supporting our observations, lithium was previously shown to overcome neonatal isolation-induced autistic-like behavior in a rat model by restoring the compromised balance between excitatory/inhibitory synaptic activities (Wu et al., 2014). Lithium was also proposed as a potential therapeutic approach for Fragile X syndrome. Chronic lithium treatment contributes to normalized behavior and synaptic transmission in mice models of Fragile X syndrome and pilot clinical trials have shown promising results in individuals with Fragile X syndrome (Liu and Smith, 2014). This suggests that lithium may have a global positive effect on glutamatergic synapse genesis and homeostasis. This would be consistent with mRNA profiling performed in the present study, which indicate that lithium positively regulates SHANK3 together with 6 of its protein-partners at the glutamatergic synapse. How lithium regulates SHANK3 remains, however, to be fully characterized as lithium acts on a large variety of signaling pathways (Wu et al., 2013, Pasquali et al., 2010, Millan, 2013). Our results suggest that lithium, as VPA, may act, in part, through its HDAC inhibitory activity and regulate SHANK3 via an epigenetic mechanism. Lithium was demonstrated to inhibit inositol dependent enzymes by depleting inositol cellular content (Gould et al., 2004). Interestingly, HDAC were recently demonstrated as depending of inositol for proper activity (Watson et al., 2016). This suggests that Lithium may directly inhibit HDAC activity via inositol depletion. In addition, we found that Lithium, as VPA, down-regulated directly histone expression. Together, lithium and VPA may regulate SHANK3 at an epigenetic levels via several pathways linked to histone linkage to DNA. Clinical assessment of patient AUN-003 suggested that lithium, initially provided for bipolar disorders, improved also her social and interaction behavior as evaluated using SRS and ADOS. Interestingly, after one year of treatment, performances on SRS and ADOS were better than when she was first enrolled in the study 2.5 years before (T0), i.e. before the beginning of mood deregulation. These effects on scales exploring ASD were not correlated with performance on global adaptation behavior scales (Vineland and GAS), suggesting that the improvement recorded for ASD symptoms was not, at least not only due to a recovery from regression or a placebo effect. While encouraging, our case report also revealed the emergence of ADHD in this patient that should serve as a cautionary tale for the premature use of this approved medication for a complex condition such as ASD. These symptoms resembled the hyperkinetic, manic-like, disorders described previously in two patients with SHANK3 duplication and mimicked in SHANK3 overexpressing mice(Han et al., 2013). In this mouse model, lithium was inefficient at correcting SHANK3 dependent maniac-like behavior, in contrast to VPA, an atypical pharmacological profile for manic-like disorders. In light of our results, the up-regulation of SHANK3 by lithium may explain the lack of efficacy. Regarding VPA, additional action of this compound on the GABAergic system may overall result in a correction of the manic-like behavior despite up-regulating SHANK3. Collectively, this suggests that lithium may have improved ASD symptoms but induced ADHD-like behavior by increasing SHANK3 in AUN-003. These observations highlight the fact that proper SHANK3 dosage is critical for normal brain function, and warrant deeper investigation of the mechanisms of action of lithium, VPA and related agents and their potential benefits for patients with SHANK3 haplo-insufficiency. Our work pleads for a deeper characterization of instrumental pathways sustaining lithium action on SHANK3 to identify more specific molecules and avoid potential non-desired side effects resulting from the inhibition of multiple cellular targets not involved in SHANK3 regulation. Additionally, animal testing will be critical to elucidate lithium efficacy. So far, the most widely studied models are mice fully knock-out for SHANK3 isoforms. These mice exhibit strong and robust abnormal behavior (review in Jiang and Ehlers, 2013). Consistently, validation of pharmacological approaches preferentially uses these KO mice (Bozdagi et al., 2013, Bidinosti et al., 2016). However, our approach is based upon the identification of compounds rescuing SHANK3 associated phenotypes by increasing SHANK3 mRNA levels. Testing of lithium would therefore need to be performed in heterozygous mice with one wild-type allele. Unfortunately, these mice develop only very specific and discrete phenotypes (Bozdagi et al., 2010), the robustness of which limiting their use as model for testing efficiency of pharmacological approaches. Finally, formal conclusions would come from a randomized double blind clinical trial against placebo that will be needed to accurately validate the effect of lithium in patients carrying SHANK3 loss of function mutations and exclude any possibility of symptom improvement solely due to change in educational care or any other environmental factors.
5. Conclusion
The main result of this study is the demonstration that lithium, the FDA-approved drug for bipolar disorders, regulates SHANK3 expression in a way that bears therapeutic potential for patients with ASD associated to SHANK3 haploinsufficiency. This result has been obtained on the basis of a drug screening challenging molecular and functional phenotypes relevant to the disease in neurons derived from pluripotent stem cell lines, including from patients awaiting a treatment. This novel methodology for drug discovery is applicable to other forms of ASD with contrasting pathophysiological substrates, as well as to other CNS disorders with known molecular anomalies. As such, it offers an avenue to the stratification of treatment and, ultimately, personalized medicine for psychiatric disorders.
The following are the supplementary data related to this article.
Primary screening results
List of tested compounds and their concentration used in the primary screening as well as SHANK3 fold changes compared to DMSO treated cells. Results are provided as mean ± SD of 4 replicates (independently treated neuronal cultures).
List of antibodies used in this study
Name, host species, working concentrations and providers of the primary antibodies used in this study.
mRNA profiling of neuronal and synaptic gene following lithium and VPA treatment
Results of fold change expression of treated cells vs DMSO for all the genes present in the two mRNA profiling assays, namely the “synaptic protein” and the “GABA Glutamate” arrays. Results are provided as mean ± SD of 3 replicates (independently treated neuronal cultures).
Transcriptomic analysis following lithium and VPA treatment
Lists of gene modified in 14 days human neurons following treatment with VPA or lithium. Results are expressed as mean fold change of 3 independently treated neuronal cultures compared to DMSO treated cultures. The lists were established using the following threshold: p value < 0.05 (GSA algorithm, Partek Inc.), Fold change > 1.5 or − 1.5 compared to DMSO. Pathway analyses were performed using Panther algorithm via the open access EnrichR on line platform.
Complete clinical assessment of patient AUN-003
Assessment scores obtained while exploring the patient AUN-003. SRS: Social Responsiveness Scale, ADOS: Autism Diagnosis Observational Scale, PPVT: Peabody Picture Vocabulary Test, RPM: Raven's progressive matrices, GAS: Global Assessment Scale, Y-MRS: Young-Mania rating Scale, ADHD: Attention Deficit Hyperactive Disorder rating scale.
Contributors
Conception and design of the study: AB, MP, TB, RD; Screening and in vitro experimental design: HD, AP, AB, LC, CB, FC, MJM; Data collection, analysis and interpretation: HD, AP, FRA, LC, CB, JP, TB, RD, AB; Compound library design and providing: HD, MJM, FC, AB; Transcriptomic studies: ALP, MJ, Patients recruitment, genetic characterization and sampling: CN, TB, RD; Patients clinical evaluation and post-treatment follow-up: FRA, RD; Report first draft writing: HD, AB; Critical review and editing, MJM, TB, MP, RD. Study supervision: AB.
Funding
Association Française contre les Myopathies (AFM-Téléthon), French National Health Institute (INSERM), French National Research Agency (ANR), Bettencourt-Schueller Foundation, Cognacq Jay Foundation, Fondamental Foundation, Servier Laboratories.
Role of the Funding Sources
Funders of the study had no role in study design, data analysis, data interpretation, or writing of the report. All authors had full access to the data and were jointly responsible for the decision to submit the manuscript.
Aknowledgements
The authors are thankful to I-Stem's HTS platform staff for constant technical support. We thank the cell bank of Pitié-Salpétrière hospital and the Clinical Investigation Center of Robert Debré hospital for assistance with patient recruitment, information, sampling and fibroblast preparation. H.D. received a PhD grant from Servier's Laboratories. I-Stem is part of the Biotherapies Institute for Rare Diseases (BIRD) supported by the Association Française contre les Myopathies (AFM-Téléthon). This study has been in part funded by grants from “Investissements d'Avenir” – (ANR-11-INBS-0009 - INGESTEM – and ANR-11-INBS-0011 - NeurATRIS), the French National Research Agency ANR (ANR-13-SAMA-0006; SynDivAutism), the Laboratory of Excellence GENMAD (ANR-10-LABX-0013), the Bettencourt-Schueller Foundation, the Cognacq Jay Foundation, and the Fondamental Foundation. This study used samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository, as well as clinical data. NINDS Repository sample numbers corresponding to the samples used are GM 1869.
References
- Bidinosti M., Botta P., Kruttner S. CLK2 inhibition ameliorates autistic features associated with SHANK3 deficiency. Science. 2016;351(6278):1199–1203. doi: 10.1126/science.aad5487. [DOI] [PubMed] [Google Scholar]
- Bittker J.A. High-throughput RT-PCR for small-molecule screening assays. Curr. Protoc. Chem. Biol. 2012;4(1):49–63. doi: 10.1002/9780470559277.ch110204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers T.M., Bockmann J., Kreutz M.R., Gundelfinger E.D. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J. Neurochem. 2002;81(5):903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- Boissart C., Nissan X., Giraud-Triboult K., Peschanski M., Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139(7):1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- Boissart C., Poulet A., Georges P. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl. Psychiatry. 2013;3:e294. doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015;16(9):551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- Bozdagi O., Sakurai T., Papapetrou D. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol. Autism. 2010;1(1):15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O., Tavassoli T., Buxbaum J.D. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol. Autism. 2013;4(1):9. doi: 10.1186/2040-2392-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Zou X., Cui Y. Rapid throughput analysis demonstrates that chemicals with distinct seizurogenic mechanisms differentially alter Ca2 + dynamics in networks formed by hippocampal neurons in culture. Mol. Pharmacol. 2015;87(4):595–605. doi: 10.1124/mol.114.096701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Huang L.D., Jiang Y.M., Manji H.K. The mood-stabilizing agent valproate inhibits the activity of glycogen synthase kinase-3. J. Neurochem. 1999;72(3):1327–1330. doi: 10.1046/j.1471-4159.2000.0721327.x. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme R., Ey E., Toro R., Leboyer M., Gillberg C., Bourgeron T. Progress toward treatments for synaptic defects in autism. Nat. Med. 2013;19(6):685–694. doi: 10.1038/nm.3193. [DOI] [PubMed] [Google Scholar]
- Dravid S.M., Murray T.F. Spontaneous synchronized calcium oscillations in neocortical neurons in the presence of physiological [Mg(2 +)]: involvement of AMPA/kainate and metabotropic glutamate receptors. Brain Res. 2004;1006(1):8–17. doi: 10.1016/j.brainres.2004.01.059. [DOI] [PubMed] [Google Scholar]
- Durand C.M., Betancur C., Boeckers T.M. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges P., Boissart C., Poulet A., Peschanski M., Benchoua A. Protein kinase-A inhibition is sufficient to support human neural stem cells self-renewal. Stem Cells. 2015 doi: 10.1002/stem.2194. [DOI] [PubMed] [Google Scholar]
- Gould T.D., Quiroz J.A., Singh J., Zarate C.A., Manji H.K. Emerging experimental therapeutics for bipolar disorder: insights from the molecular and cellular actions of current mood stabilizers. Mol. Psychiatry. 2004;9(8):734–755. doi: 10.1038/sj.mp.4001518. [DOI] [PubMed] [Google Scholar]
- Grabrucker A.M., Knight M.J., Proepper C. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011;30(3):569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Holder J.L., Jr., Schaaf C.P. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503(7474):72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.H., Ehlers M.D. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78(1):8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.A., Christian S.L. Genetics of autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 2009;9(3):188–197. doi: 10.1007/s11910-009-0029-2. [DOI] [PubMed] [Google Scholar]
- Leblond C.S., Nava C., Polge A. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 2014;10(9):e1004580. doi: 10.1371/journal.pgen.1004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.S., Pirooznia M., Guintivano J. Search for common targets of lithium and valproic acid identifies novel epigenetic effects of lithium on the rat leptin receptor gene. Transl. Psychiatry. 2015;5:e600. doi: 10.1038/tp.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Kida Y.S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Smith C.B. Lithium: a promising treatment for fragile X syndrome. ACS Chem. Neurosci. 2014;5(6):477–483. doi: 10.1021/cn500077p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Monteiro P., Zhou Y. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530(7591):481–484. doi: 10.1038/nature16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan M.J. An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology. 2013;68:2–82. doi: 10.1016/j.neuropharm.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Newschaffer C.J., Croen L.A., Daniels J. The epidemiology of autism spectrum disorders. Annu. Rev. Public Health. 2007;28:235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- Pasquali L., Busceti C.L., Fulceri F., Paparelli A., Fornai F. Intracellular pathways underlying the effects of lithium. Behav. Pharmacol. 2010;21(5–6):473–492. doi: 10.1097/FBP.0b013e32833da5da. [DOI] [PubMed] [Google Scholar]
- Peca J., Feliciano C., Ting J.T. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M., Stelzer Y., Bar-Nur O., Mayshar Y., Eden A., Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27(11):2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- Sehgal P.B., Darnell J.E., Jr., Tamm I. The inhibition by DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976;9(3):473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- Shcheglovitov A., Shcheglovitova O., Yazawa M. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503(7475):267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooren W., Lindemann L., Ghosh A., Santarelli L. Synapse dysfunction in autism: a molecular medicine approach to drug discovery in neurodevelopmental disorders. Trends Pharmacol. Sci. 2012;33(12):669–684. doi: 10.1016/j.tips.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Toro R., Konyukh M., Delorme R. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26(8):363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Urbach A., Bar-Nur O., Daley G.Q., Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6(5):407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.J., Millard C.J., Riley A.M. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 2016;7:11262. doi: 10.1038/ncomms11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Bai Y., Tan T. Lithium ameliorates autistic-like behaviors induced by neonatal isolation in rats. Front. Behav. Neurosci. 2014;8:234. doi: 10.3389/fnbeh.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Zheng S.D., Huang H.L. Lithium down-regulates histone deacetylase 1 (HDAC1) and induces degradation of mutant huntingtin. J. Biol. Chem. 2013;288(49):35500–35510. doi: 10.1074/jbc.M113.479865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Danko T., Botelho S.C. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science. 2016 doi: 10.1126/science.aaf2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primary screening results
List of tested compounds and their concentration used in the primary screening as well as SHANK3 fold changes compared to DMSO treated cells. Results are provided as mean ± SD of 4 replicates (independently treated neuronal cultures).
List of antibodies used in this study
Name, host species, working concentrations and providers of the primary antibodies used in this study.
mRNA profiling of neuronal and synaptic gene following lithium and VPA treatment
Results of fold change expression of treated cells vs DMSO for all the genes present in the two mRNA profiling assays, namely the “synaptic protein” and the “GABA Glutamate” arrays. Results are provided as mean ± SD of 3 replicates (independently treated neuronal cultures).
Transcriptomic analysis following lithium and VPA treatment
Lists of gene modified in 14 days human neurons following treatment with VPA or lithium. Results are expressed as mean fold change of 3 independently treated neuronal cultures compared to DMSO treated cultures. The lists were established using the following threshold: p value < 0.05 (GSA algorithm, Partek Inc.), Fold change > 1.5 or − 1.5 compared to DMSO. Pathway analyses were performed using Panther algorithm via the open access EnrichR on line platform.
Complete clinical assessment of patient AUN-003
Assessment scores obtained while exploring the patient AUN-003. SRS: Social Responsiveness Scale, ADOS: Autism Diagnosis Observational Scale, PPVT: Peabody Picture Vocabulary Test, RPM: Raven's progressive matrices, GAS: Global Assessment Scale, Y-MRS: Young-Mania rating Scale, ADHD: Attention Deficit Hyperactive Disorder rating scale.