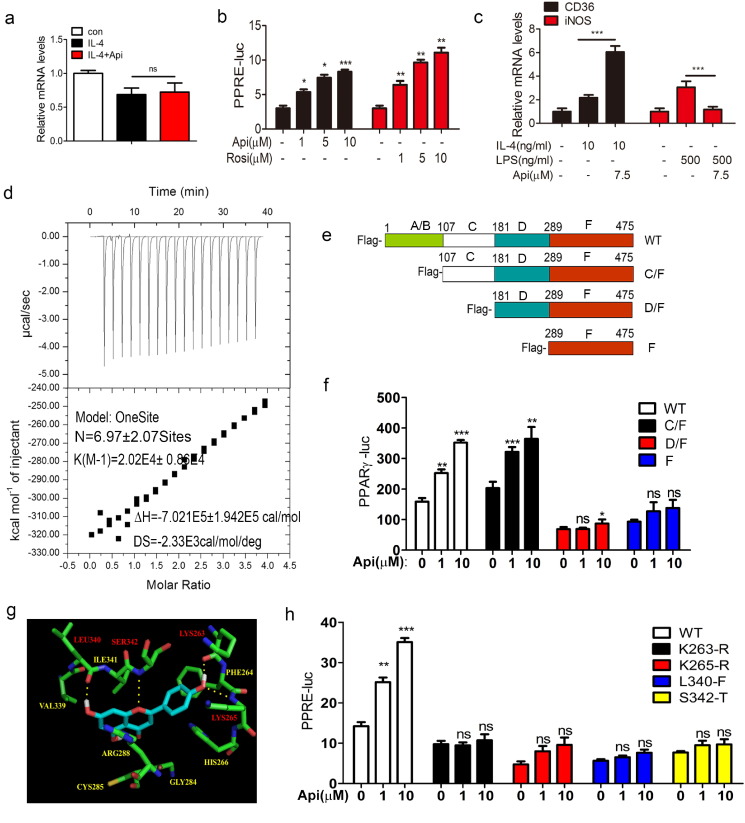
Fig. 4.
Api binds to and activates PPARγ. (a) ANA-1 macrophages was treated with 10 ng/mL IL-4 or 10 ng/mL IL-4 plus 7.5 μM Api simultaneously for 24 h, the mRNA level PPARγ was measured by using qRT-PCR. Data indicate fold induction (normalized by β-actin signal). All values are expressed as mean ± SEM. Statistical analysis is based on one-way ANOVA followed by a Dunnett's test. ns, no significant difference. (b) Transcriptional activation of PPARγ in cells treated with the indicated dosages of Api or Rosi. HER293T cells were transfected with pIRES-mPPARγ/PPRE and pRL-control using Lipofectamine2000. Then cells were pre-treated with apigenin for 24 h. Luciferase activities were measured by using the dual-luciferase reporter assay system. All values are expressed as mean ± SEM. Statistical analysis is based on one-way ANOVA followed by a Dunnett's test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with 0 group. (c) ANA-1 macrophages was treated with the indicated group for 24 h, the mRNA level PPARγ activation related genes CD36 and iNOS was measured by using qRT-PCR. Data indicate fold induction (normalized by β-actin signal). All values are expressed as mean ± SEM. Statistical analysis is based on one-way ANOVA followed by a Dunnett's test. ns, no significant difference. (d) ITC data for binding of Api to PPARγ. The upper panels show the raw data, and the lower panels show the corresponding binding isotherm fitted according to the “one binding site” model. Reference titration of ligand into buffer was used to correct for heat of dilution. The thermodynamic parameters (K, ΔH, and ΔS) are indicated under the below. (e) The deletion mutant model. (f) HER293T cells were transfected with pIRES-mPPARγ truncated mutants/PPRE and pRL-control using Lipofectamine2000. Then cells were pre-treated with Api (1 μM, 10 μM) for 24 h. Luciferase activities were measured by using the dual-luciferase reporter assay system. All values are expressed as mean ± SEM. Statistical analysis is based on one-way ANOVA followed by a Dunnett's test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with 0 group. (g) Auto dock model of Api binding to the PPARγ. Hydrogen bonding was built between Api and the Lys263, Lys265, Leu340 and Ser 342 sites of PPARγ. (h) Mutants disturbed Api from activating PPARγ analyzed using the dual-luciferase reporter assay system. HER293T cells were transfected with pIRES-mPPARγ point mutants/PPRE and pRL-control using Lipofectamine2000. Then cells were pre-treated with Api (1 μM, 10 μM) for 24 h. Luciferase activities were measured by using the dual-luciferase reporter assay system. All values are expressed as mean ± SEM. Statistical analysis is based on one-way ANOVA followed by a Dunnett's test. *P < 0.05, **P < 0.01, ***P < 0.001 compared with 0 group.