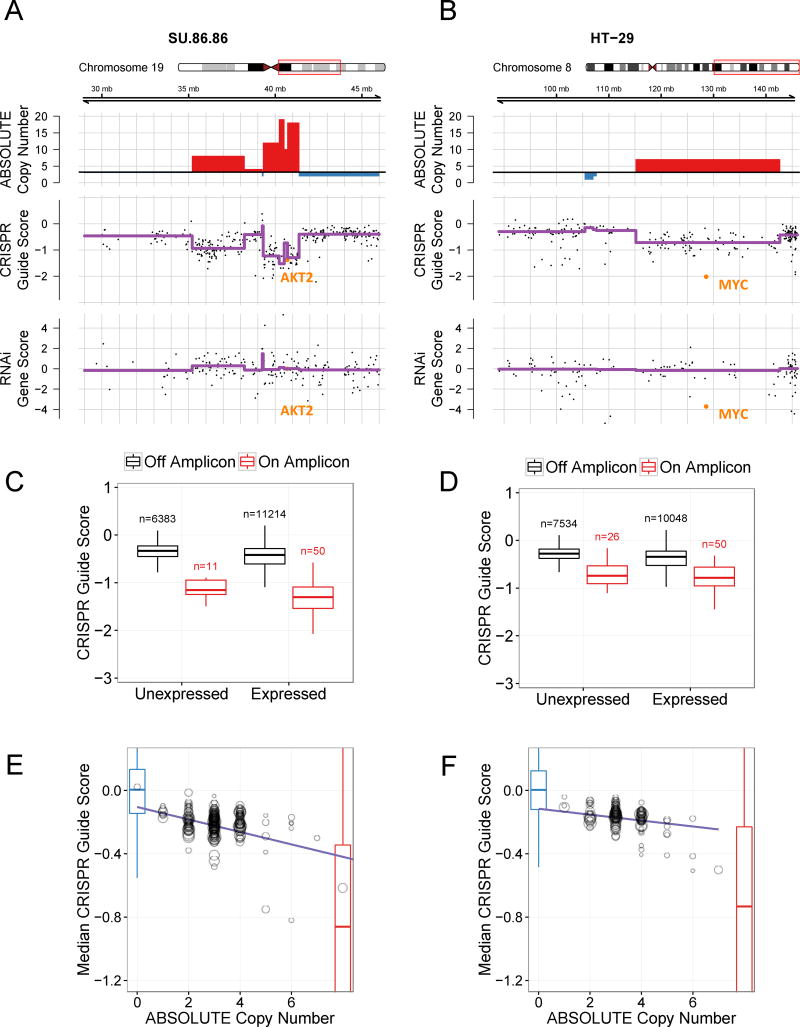
Figure 2.
Genome scale CRISPR-Cas9 screening identifies a strong correlation between copy number and sensitivity to CRISPR-Cas9 genome editing. Two cell lines are shown: SU86.86 (A, C, E) and HT29 (B, D, F). (A) Chromosome 19q amplicon in SU86.86 and (B) chromosome 8q amplicon in HT29: Three tracks are plotted along genomic coordinates within the region defined the red box on the chromosome schematic. Top: ABSOLUTE genomic copy number from Cancer Cell Line Encyclopedia (CCLE) SNP arrays with red indicating copy number gain above average ploidy and blue indicating copy number loss below average ploidy; Middle: CRISPR-Cas9 guide scores plotted according to the 2nd most dependent sgRNA for each gene with purple trend line indicating the mean CRISPR guide score for each copy number segment defined from the above track; Bottom: RNAi gene dependency scores. AKT2 and MYC, known driver oncogenes at these loci, respectively, are highlighted in orange. For RNAi data, shRNAs targeting AKT2 used in Project Achilles were not effective in suppressing AKT2. (C, D) Boxplots of CRISPR guide scores for both expressed and not expressed genes located on (red) or off (black) of the chromosome 19q amplicon in SU86.86 (C) and the chromosome 8q amplicon in HT29 (D). For the SU86.86, the amplicon represented in panel C red box plots ranges from 39.3–41.4 Mb on the corresponding plot in panel A. The number of represented genes is noted above each box plot. (E, F) For each copy-number-defined genomic segment, median CRISPR-Cas9 guide score is plotted against copy number. Each circle represents a single genomic segment of defined copy number for the indicated cell line. The size of the circle corresponds to the number of sgRNAs targeting that segment. Non-targeting negative control sgRNAs are shown with a blue boxplot and known cell essential genes (defined as positive controls) are shown as a red boxplot embedded within the plot.