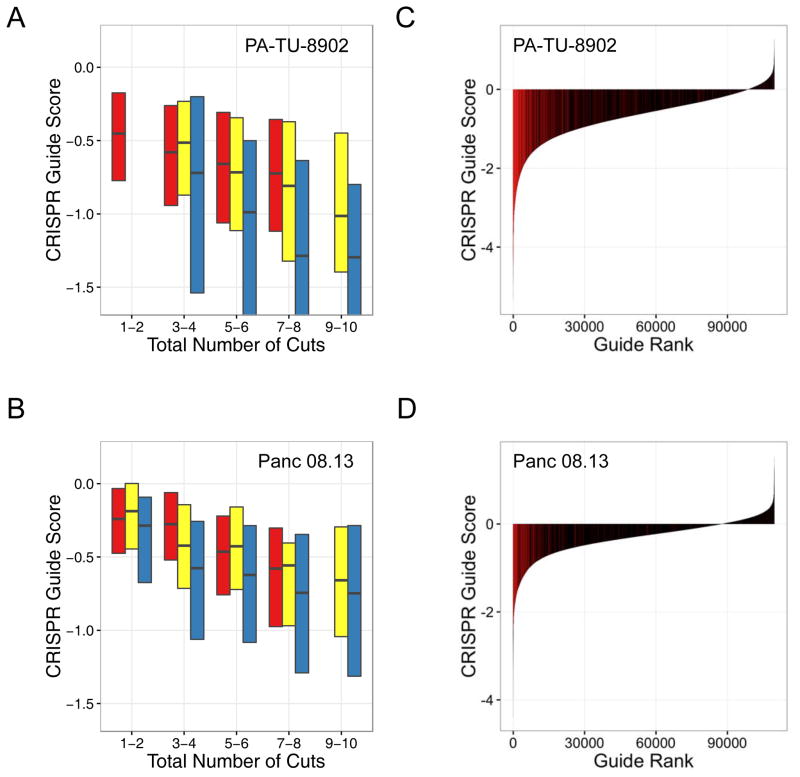
Figure 5.
sgRNAs targeting multiple chromosomes show greater sensitivity to CRISPR-Cas9-induced cutting. Data from two representative cell lines are shown (PA-TU-8902, A, C; Panc 08.13, B, D) (A, B) Boxplots of CRISPR-Cas9 sensitivity to the predicted number of CRISPR-Cas9-induced DNA cuts. CRISPR-Cas9 guide scores are shown on the Y-axis and the predicted number of DNA cuts is shown on the X-axis. sgRNAs are divided into three groups. In red are sgRNAs that target a single locus, and therefore total number of predicted cuts is based on copy number. In yellow are sgRNAs that target multiple loci within a single chromosome (intra-chromosomal). In blue are sgRNAs that target multiple loci across multiple chromosomes (inter-chromosomal). The analysis demonstrates a more potent detrimental influence on cell viability for multiple CRISPR-Cas9-induced DNA cuts across multiple chromosomes (inter-chromosomal) as compared to those restricted to a single chromosome (intra-chromosomal). Multiple linear regression accounting for difference in total number of cuts for inter-chromosomal vs. intra-chromosomal: panel A, β = −0.27, p = 2.64e-22; panel C, β = −0.16, p = 4.97e-22. (C, D) Waterfall plots showing CRISPR guide scores for all sgRNAs in the pooled screens performed on the indicated cell lines. sgRNAs from the multiple alignment analysis targeting multiple chromosomes with >10 predicted target sites are shown in red and are significantly enriched with negative CRISPR-Cas9 guide scores relative to all other sgRNAs in the library (one-sided Kolmogorov–Smirnov test: p = 2.13E-159, C; p = 9.17E-88, D) These data highlight the potent detrimental effect that these sgRNAs have on cell proliferation and viability within the screen.