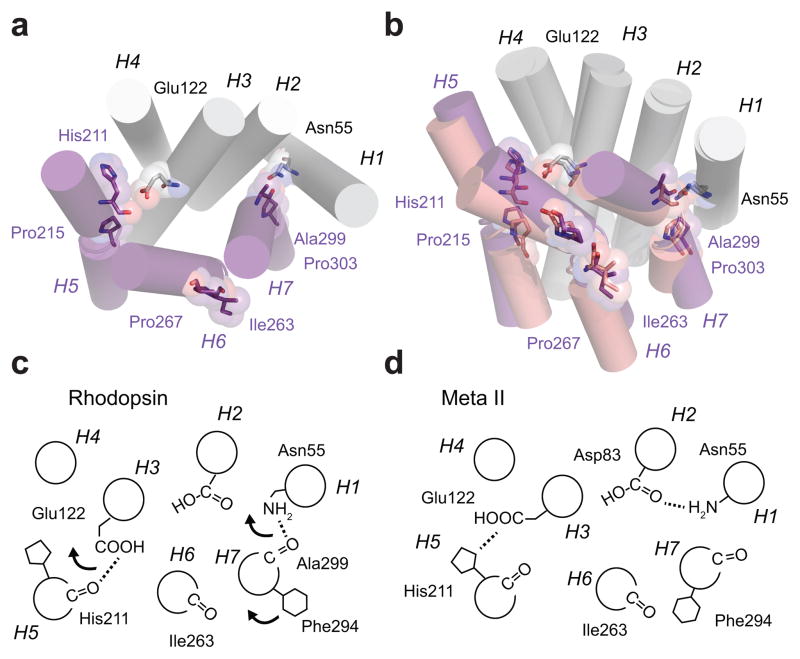
Figure 4.
Receptor activation leads to repacking of helices H5–H7 on the TM core of rhodopsin. (a) Inactive structure of rhodopsin. Helices H1–H4 form a scaffold onto which helices H5–H7 pack. The key packing contacts on H5 and H7 are associated with Pro2155.50 and Pro3037.50 and their corresponding i-4 carbonyls. Retinal isomerization disrupts both interactions. For H5, the β-ionone ring has a steric clash at the position of the His2115.46-Glu1223.37 hydrogen bond upon conversion the all-trans configuration. For H7, the retinal is covalently attached to Lys2967.43. Upon isomerization, Trp2656.48 on H6 rotates away from H7 and disrupts a water mediated hydrogen bond with Asn3027.49, which is part of a hydrogen bonding network stretching from Asn551.50 and Asp832.50 to the Ala2997.46 C=O. (b) Overlap of the crystal structures of rhodopsin (1GZM10, purple) and Meta II (3PQR13, light purple) showing the positions of the TM helices. The disruption of the interactions of the His2115.46 C=O and Ala2997.46 C=O with the H1–H4 scaffold allows helices H5–H7 to reorient. (c,d) Schematic of the hydrogen bonding changes occurring between inactive rhodopsin and active Meta II rhodopsin.