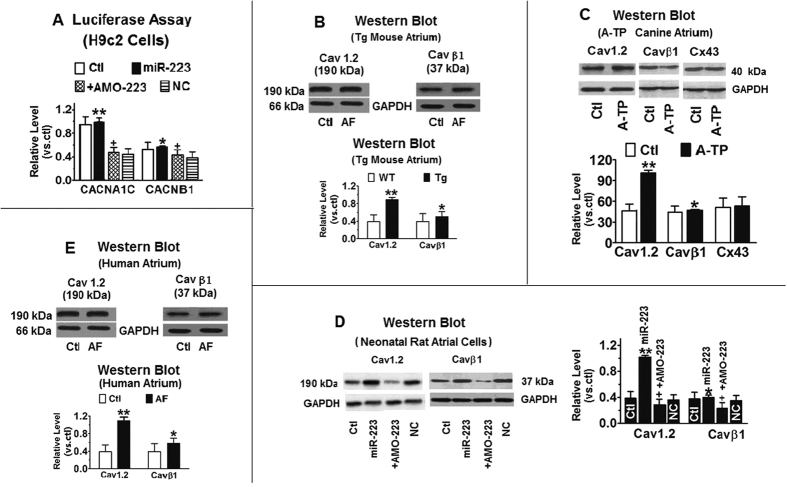
Figure 4. CACNA1C/Cav1.2 as the target gene for miR-223.
(A) Luciferase assays in H9c2 cells for posttranscriptional excitation of CACNA1C (the gene encoding cardiac L-type Ca2+ channel αlc (Cav1.2) protein). Regulation of Cav1.2 protein expression by miR-223. Western blot showing the effect of miR-223 (n = 10) and antisense miR-223 (AMO-223; n = 10) on Cav1.2 protein expression in H9c2 rat atrial cells. MiR-223 (10 nmol/L) and AMO-223 (10 nmol/L) were transfected, and then the cells treated with ox-LDL. Ctl, mock-treated with ox-LDL; miR-NC, NC miRNA. **P < 0.001, *P < 0.05 vs. NC; **P > 0.05, *P > 0.05 vs. Ctl; +P < 0.05 vs. NC; +P < 0.001 vs. Ctl; (B) Western blot analysis of Cav1.2 and Cav β1 proteins in atrial tissues from miR-223 Tg and WT mice. **P < 0.001 vs WT, unpaired t test; n = 10 mice in each group. *P < 0.05 vs. Ctl. (C) Western blot analysis of Cav1.2 and Cav β1 proteins in atrial tissues from A-TP dogs. Connexin 43 protein (Cx43) was used as the NC. **P < 0.001 vs. Ctl, unpaired t test; n = 10 protein samples from two dogs in each group. *P < 0.05 vs. Ctl. (D) Effect of miR-223 on Cav1.2 and Cav β1 transcript levels in miR-223/Tg mice and cultured neonatal rat atrial myocytes. n = 10 RNA samples in each group. **P < 0.001 vs Ctl, *P < 0.05 vs. Ctl, unpaired t test. (E) Cav1.2 and Cav β1 protein upregulation in atrial tissues from AF patients with rheumatic heart disease compared with non-AF subjects (Ctl). **P < 0.001 vs Ctl, unpaired t test; n = 20 patients in each group. *P < 0.05 vs. Ctl.