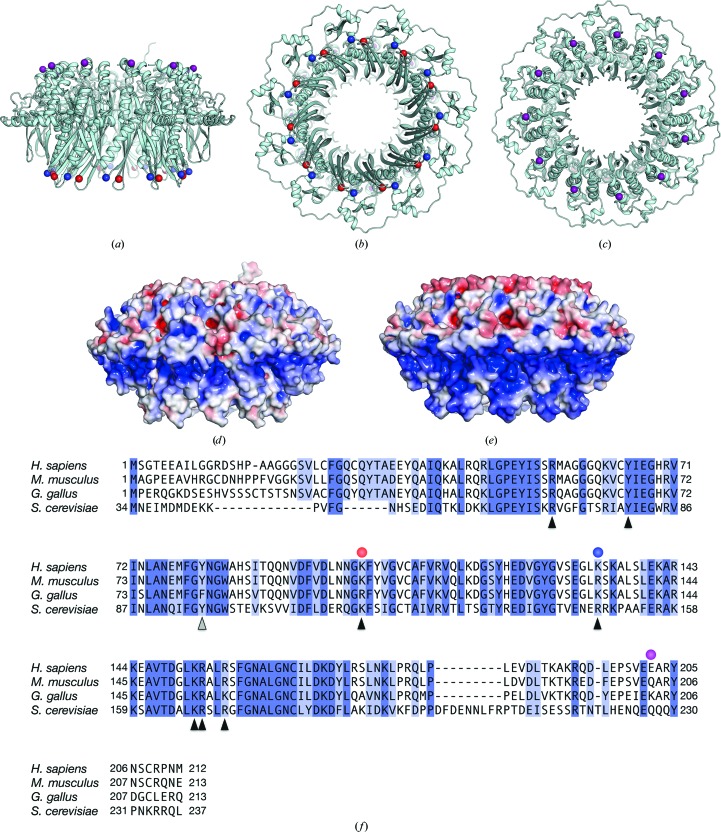
Figure 1.
Crystal structure of the human RAD521–212 K102A/K133A/E202A protein. The mutant RAD521–212 structure is depicted as a ribbon representation, and its side (a), bottom (b) and top (c) views are shown. Amino-acid positions 102, 133 and 202 (coloured red, blue and purple, respectively) are shown as spheres. (d) A surface representation of the mutant RAD521–212 structure, viewed from the side [the same view as in (a)]. The surface is coloured according to the local electrostatic potential, from −5.0k B T (red) to +5.0k B T (blue). (e) A surface representation of nonmutated RAD521–212, coloured similarly to the mutant RAD521–212 shown in (d). (f) Amino-acid sequence comparison of human, mouse, chicken and yeast (Saccharomyces cerevisiae) RAD52 proteins. Amino-acid positions 102, 133 and 202 (coloured red, blue and purple, respectively) are indicated by circles. The amino-acid residues of human RAD52 that are important for DNA binding and self-association are indicated by black and grey triangles, respectively.