Abstract
RNA structure is intimately related to function, yet methods to identify base-paired RNA strands in a transcriptome-wide manner in cells have remained elusive. One recent paper in Cell and two in Molecular Cell describe related methods to identify RNA sequences that interact in living cells, setting the stage for break-throughs in our understanding of RNA structure and function.
Just like proteins, RNA molecules adopt 3D structures that are important for their processing and function. Although it is well known that several classes of RNAs such as transfer RNA (tRNA), ribosomal RNA (rRNA), and small nuclear RNAs (snRNAs) are highly structured, we know remarkably little about the structures of most RNA molecules encoded in our genomes. In fact, nearly all textbooks depict messenger RNAs as linear, non-structured, single-stranded molecules. Not only do most RNAs likely have specific 3D structures, but these structures are almost certainly important for the functions of these RNAs.
Over the past several decades, a large toolbox of diverse methods has been developed to characterize RNA secondary structures and identify RNA sequences that base-pair with one another, whether in cis or in trans. These include computational modeling, phylogenetic analysis, genetic testing, and biochemical probing (reviewed in Wan et al., 2011). For example, several computational tools exist that use thermodynamic rules to predict RNA secondary structure based on the primary sequence. Comparative analysis of orthologous sequences from multiple species can also provide evidence supporting RNA secondary structures. Specifically, co-variation of distantly located bases is indicative that those nucleotides base-pair with one another. Similarly, unpaired sequences often exhibit more rapid and less-constrained sequence divergence than base-paired sequences. Traditional genetic experimentation can be used to test and validate secondary structure predictions. In fact, much of our knowledge of the transient and dynamic base-pairing within and between snRNAs involved in pre-mRNA splicing was either identified or proven by genetic experiments in yeast by generating mutations predicted to disrupt a structure and corresponding compensatory mutations that restore the structure. Chemicals and enzymes provide additional means of probing RNA secondary structure. For example, sequence- or structure-specific ribonucleases can be used to identify paired and unpaired regions of specific RNAs. Similar information can also be obtained using chemicals that modify RNA, such as dimethyl sulfate (DMS), which methylates unpaired A and C bases, creating adducts that reverse transcriptase cannot bypass. The chemical and enzymatic probing methods have typically been restricted to use with individual in vitro transcribed RNA. As a result, these approaches may not accurately reflect the structures that RNAs adopt in vivo and, until recently, could not be used to simultaneously study multiple RNAs.
To address these limitations, several groups have adapted these methods by coupling them to high-throughput sequencing, allowing for RNA structures to be investigated on a large scale and even transcriptome-wide manner (Table 1). For example, FragSeq (Underwood et al., 2010), PARS (Kertesz et al., 2010), and SHAPE-seq (Lucks et al., 2011) use P1 nuclease, RNase V1 and S1 nuclease, and 1-methyl-7-nitroisatoic anhydride (1M7), respectively, to probe the structures of a large pool of synthetic RNAs or total RNA after extraction from cells and provided information on the conformation RNAs adopted in vitro. These approaches were followed by the development of mod-seq (Talkish et al., 2014), DMS-seq (Rouskin et al., 2014), Structure-seq (Ding et al., 2014), icSHAPE (Spitale et al., 2015), and SHAPE-Map (Smola et al., 2015), which all perform the conformational probing in cells prior to purification of the RNA and therefore provide information on the structures of RNA as they were in living cells. While these methods represented important advances and provided specific information about which regions of an RNA were single- or double-stranded in cells, none of them were able to identify which RNA sequences were directly pairing with one another. As a result, until now, methods to resolve long-range structures, alternative RNA conformations, pseudoknots, and RNAs that interact in trans, especially on a transcriptome-wide scale, have not existed.
Table 1.
Transcriptome-wide RNA Structure Probing Methods
| Assay | Probing Agent | Detection | In Vitro Probing | In Vivo Probing | Reference |
|---|---|---|---|---|---|
| FragSeq | P1 nuclease | single-stranded bases | X | Underwood et al., 2010 | |
| PARS | RNase V1 and S1 nuclease |
paired and single- stranded regions |
X | Kertesz et al., 2010 | |
| SHAPE-seq | 1M7 | single-stranded bases | X | Lucks et al., 2011 | |
| mod-seq | DMS | unpaired A & C | X | Talkish et al., 2014 | |
| DMS-seq | DMS | unpaired A & C | X | X | Rouskin et al., 2014 |
| Structure-seq | DMS | unpaired A & C | X | X | Ding et al., 2014 |
| icSHAPE | NAI-N3 | single-stranded bases | X | Spitale et al., 2015 | |
| SHAPE-MaP | 1M7 | single-stranded or unbound bases |
X | X | Smola et al., 2015 |
| PARIS | AMT | base-paired sequence partners |
X | Lu et al., 2016 | |
| LIGR-seq | AMT | base-paired sequence partners |
X | Sharma et al., 2016 | |
| SPLASH | biotinylated psoralen | base-paired sequence partners |
X | Aw et al., 2016 |
New Methods to Identify RNA Pairs In Vivo
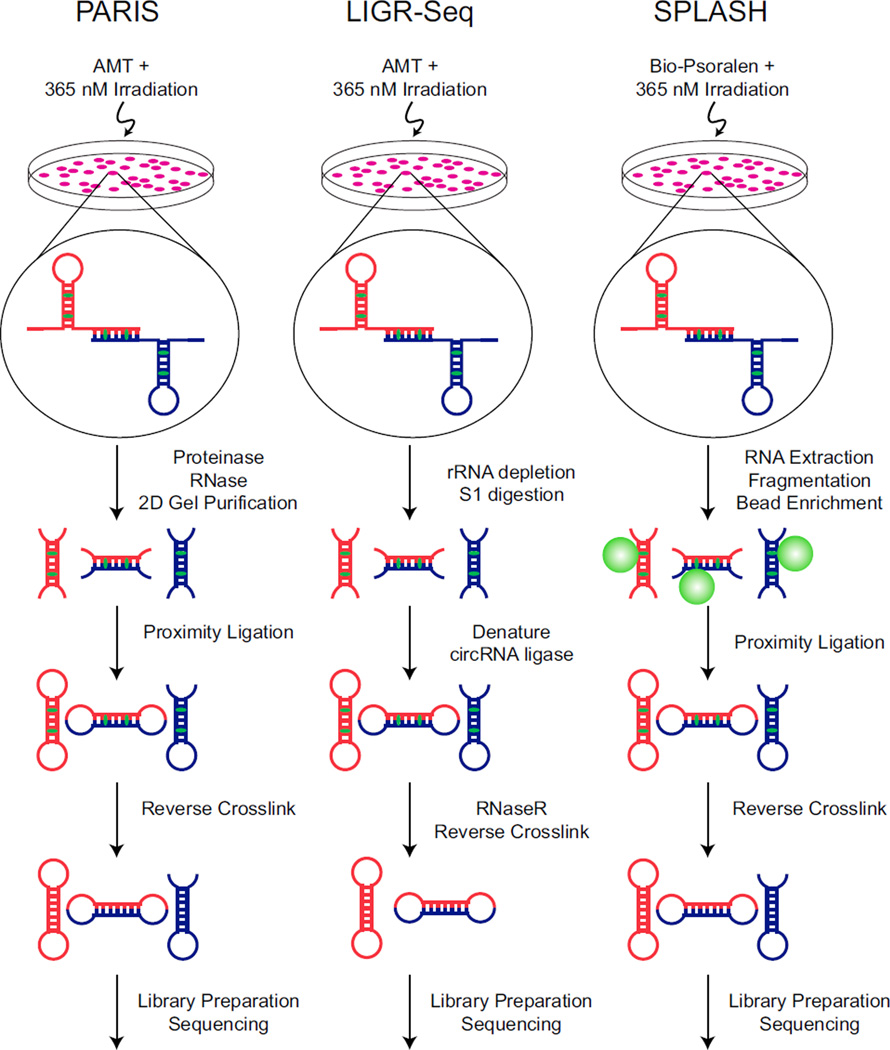
The three new studies all describe different flavors of the same general technique to capture both strands of RNA duplexes in cells and identify the sequence pairs by high-throughput sequencing (Figure 1). The three methods are called PARIS (Psoralen Analysis of RNA Interactions and Structures) (Lu et al., 2016), SPLASH (Sequencing of Psoralen crosslinked, Ligated, and Selected Hybrids) (Aw et al., 2016), and LIGR-seq (LIGation of interacting RNA followed by high-throughput Sequencing) (Sharma et al., 2016). Despite their disparate names, each protocol involves incubating cells with psoralen, crosslinking the base-paired RNAs to psoralen by UV irradiation, isolating and digesting the RNA, performing proximity ligation to link the two RNA strands together, reversing the psoralen crosslinks, sequencing the ligated fragments, and performing computational analysis to identify paired RNA sequences. However, each method employs different strategies to enrich for cross-linked RNA species.
Figure 1. Comparison of New RNA Structure Probing Methods.
Schematic overviews of the protocols for PARIS, LIGR-seq, and SPLASH are depicted. The red and blue RNA strands are meant to indicate two different RNA molecules and that the methods can be used to identify both intra- and inter-molecular interactions. AMT and Bio-Psoralen are depicted in green intercalated between the RNA base pairs.
The PARIS method (Lu et al., 2016) takes advantage of the fact that crosslinked molecules migrate anomalously in 2D polyacrylamide gels. In these experiments, non-crosslinked molecules migrate along the diagonal in the 2D gel, while crosslinked molecules migrate in an arc above the diagonal. Gel purification of RNAs migrating above the diagonal strongly enriches for crosslinked molecules, which are then used for proximity ligation and high-throughput sequencing. In LIGR-seq (Sharma et al., 2016), after crosslinking, the RNA is subjected to limiting S1 nuclease digestion and then incubated with circRNA ligase to link adjacent RNA fragments. Next, the 3′ to 5′ exoribonuclease RNase R is used to digest uncrosslinked RNAs, thereby enriching RNA molecules that were in close approximation. The authors also prepared LIGR-seq libraries from uncrosslinked and unligated samples in parallel to identify artifacts generated during library preparation. SPLASH (Aw et al., 2016) employs biotinylated psoralen, which allows crosslinked RNAs to be enriched using streptavidin affinity purification before undergoing proximity ligation. The data generated by each of these protocols yield two different sequences that could be derived from RNA fragments either from the same RNA molecule, indicating an intra-molecular base-pairing interaction, or from two different RNA molecules, indicating an inter-molecular base-pairing interaction.
To demonstrate that these methods identify biologically relevant RNA structures, each group analyzed their respective data in the context of several well-characterized intra- and inter-molecular RNA structures. For example, both Lu et al. (2016) and Sharma et al. (2016) showed that their approaches accurately detected the known interactions involving the spliceosomal snRNAs including the intra-molecular base pairing within U4 and U6 snRNAs and the inter-molecular base-pairing between U4 and U6. These methods also confirmed the inter-molecular interactions of scores of snoRNAs with their known snRNA and rRNA targets. Moreover, these approaches could also detect more complex structures such as pseudoknots. For example, PARIS detected the known pseudoknot within the RNA component of human telomerase.
More importantly than confirming known interactions, these three methods have already provided new insight into RNA interactions and RNA biology. Surprisingly, PARIS revealed that most RNAs have multiple alternative structures whose formation is mutually exclusive and that most mRNAs have long-range interactions within and between 5′ and 3′ UTRs and CDS. Moreover, many of these interactions within mRNAs are conserved between human and mouse. All three groups also identified novel inter-molecular targets of orphan snoRNAs—a group of snoRNAs for which no target RNA was previously known. Surprisingly, these snoRNA targets are not restricted to snRNAs and rRNAs, but several include mRNAs, indicating that snoRNA-directed RNA modifications may play an important role in the biogenesis and/or function of these mRNAs.
While these new RNA structure probing methods represent a truly significant advance, they provide only one aspect of the story about how RNA molecules are packaged within cells to fulfill their function. One thing that is clear from these new methods is that a more detailed view of RNA structure is obtained when combining these pairing data with more general structure probing datasets (e.g., icSHAPE) as well as evolutionary analysis. It should also be kept in mind that not only do RNAs fold into specific secondary and tertiary structures, but most RNAs are associated with RNA binding proteins in vivo. An exciting avenue of future analysis will be integrating large-scale RNA-protein interaction datasets, such as those being generated in the ENCODE project (Sundararaman et al., 2016; Van Nostrand et al., 2016), with these RNA structure datasets. Such analyses may be able to provide insight into the packaging and structure of RNPs in eukaryotic cells and therefore how RNP structure may impact function.
REFERENCES
- Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AYL, et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell. 2016;62:603–617. doi: 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl. Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. Global Mapping of Human RNA-RNA Interactions. Mol. Cell. 2016;62:618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman B, Zhan L, Blue SM, Stanton R, Elkins K, Olson S, Wei X, Van Nostrand EL, Pratt GA, Huelga SC, et al. Resources for the Comprehensive Discovery of Functional RNA Elements. Mol. Cell. 2016;61:903–913. doi: 10.1016/j.molcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, May G, Lin Y, Woolford JL, Jr, McManus CJ. Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA. 2014;20:713–720. doi: 10.1261/rna.042218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat. Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]