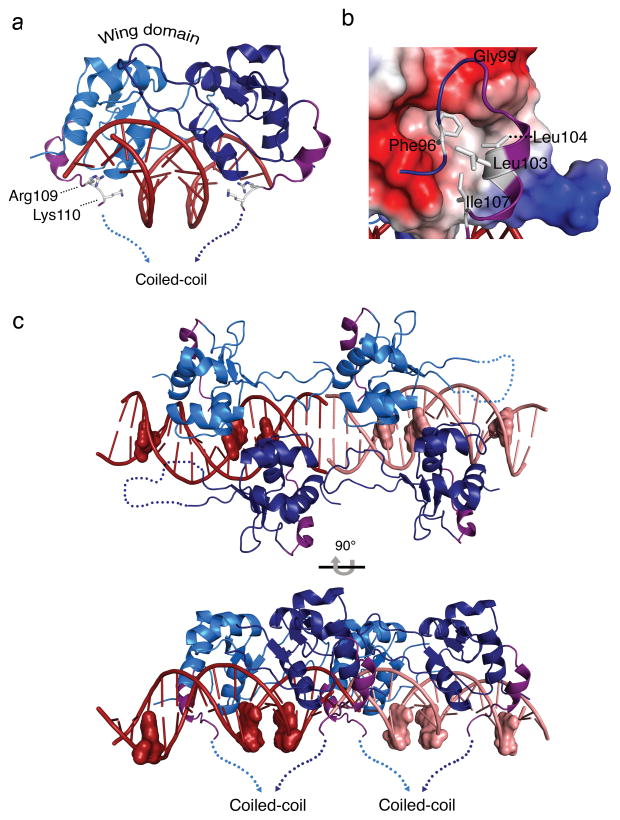
Figure 3. An HSF2 DBD carboxyl-terminal helix directs the coiled-coil multimerization domain to wrap around HSE DNA.
a) Side view of the HSF2-DBD depicting a newly resolved carboxyl-terminal helix (purple) orients the polypeptide to the underside of HSE binding site DNA, where Arg109 and Lys110 engage in direct and water-mediated DNA phosphate backbone contacts in the major groove (see Fig. S3). b) Electrostatic surface representation of the HSF2 DBD and the carboxyl-terminal helix. Red and blue represent negative and positively charged regions, respectively, and white depicts hydrophobic surfaces. The potential contours are shown on a scale from 63.41 (blue) to −63.41 kbT e−1 (red) with white representing no charge. The carboxyl-terminal helix (purple) contains two leucines (Leu103, Leu104) and an isoleucine (Ile107) that together with Phe96, pack against the hydrophobic groove presented by the core of the DBD. c) Representation of the asymmetric unit of the HSF2 DBD 3-site HSE structure. HSF2 crystallized as a dimer of dimers bound to adjacent HSE sequences shown in dark red and pink. The guanine of each nGAAn motif is represented as a surface model to show the last guanine in each HSE as unoccupied. The carboxyl-terminal helices (purple) of all four monomers direct the coiled-coil domain to the underside of the DNA, exposing the top portion of the DBD.