ABSTRACT
Ependymal cells are multiciliated cells located in the wall of the lateral ventricles of the adult mammalian brain and are key components of the subependymal zone niche, where adult neural stem cells reside. Through the movement of their motile cilia, ependymal cells control the cerebrospinal fluid flow within the ventricular system from which they receive secreted molecules and morphogens controlling self-renewal and differentiation decisions of adult neural stem cells. Multiciliated ependymal cells become fully differentiated at postnatal stages however they are specified during mid to late embryogenesis from a population of radial glial cells. Here we discuss recent findings suggesting that 2 novel molecules, Mcidas and GemC1/Lynkeas are key players on radial glial specification to ependymal cells. Both proteins were initially described as cell cycle regulators revealing sequence similarity to Geminin. They are expressed in radial glial cells committed to the ependymal cell lineage during embryogenesis, while overexpression and knock down experiments showed that are sufficient and necessary for ependymal cell generation. We propose that Mcidas and GemC1/Lynkeas are key components of the molecular cascade that promotes radial glial cells fate commitment toward multiciliated ependymal cell lineage operating upstream of c-Myb and FoxJ1.
KEYWORDS: adult neurogenic niche, fate commitment decisions, GemC1, Idas, Mcidas, multiciliated ependymal cells, multicilin, radial glial cells
Geminin superfamily is a group of proteins, which consists of Geminin and the 2 recently identified molecules named Idas and Lynkeas (also known as Mcidas and GemC1, respectively). They acquire their names after Idas and Lynkeas, twin brothers and cousins of the Gemini, Castor and Polythefkis, heroes of the ancient Greek mythology. Idas and Lynkeas were sons of Afareas and Arinis and participated in the Argonautic expedition, however they are mainly known for their implication in the death of Gemini. According to the myth Idas and Lynkeas together with the Gemini organized an animal stealing in Arcadia, but when they tried to share the loot, Idas deceived Gemini and took all the animals. After that, Gemini dissatisfied with Idas behavior left, but later they attacked to Messinia and took back the loot. Then they ambush and waited for Idas and Lynkeas. However Lynkeas saw Castor hidden in a tree and informed Idas, who killed Castor with his lance but at the same time Polythefkis killed Lynkeas. Idas then grabbed a large stone and crushed Polytheftis with it, ending up killing him. Idas was also eventually killed by Zeus, father of the Gemini, who thundered Idas after seeing his actions.34
All three members of Geminin superfamily regulate cell cycle progression and reveal similarity in their coiled-coil domain, while they interact with each other revealing high affinity in forming heretodimers.1-3 Geminin is an inhibitor of pre-replicative complex formation, Mcidas regulates DNA replication and cell cycle progression through its binding to Geminin, while GemC1 mediates pre-initiation complex formation through its interactions with TopBP1 which results in the recruitment of Cdc45 and the initiation of replication.1-6 Despite their role in DNA replication and cell cycle progression, all members of Geminin superfamily are involved on the regulation of cell fate decisions of stem cells. We and others have shown that Geminin controls self-renewal and fate commitment decisions,7-9 in several populations of stem cells through interactions with chromatin remodelling/modifying complexes, suggesting that Geminin is involved in a global epigenetic regulation of crucial transcription factors.5,9-12 We have also recently shown that Mcidas regulates the differentiation of multiciliated ependymal cells in mouse brain.13 Moreover, we have shown that GemC1/Lynkeas is key component in neural stem cells fate restriction decisions determining the generation of ependymal cells in the subependymal zone (SEZ).13
Our findings regarding the implication of Mcidas and GemC1/Lynkeas in ependymal cells differentiation13 is of great importance as they propose how the molecular cascade, responsible for the differentiation of multiciliated ependymal cells, is initiated. Ependymal cells are non-proliferating multiciliated cells located in the adult SEZ niche in the wall of the lateral ventricles, one of the 2 main neurogenic zones in adult mammalian brain where new neurons continue to be generated throughout adulthood.14,15 The main neural stem cell type in the SEZ niche responsible for the generation of neurons is type-B1 neural stem cells, which are astroglial-like cells. Type-B1 cells are mainly quiescent, but upon specific stimuli they are activated and perform self-renewal and differentiation divisions with final purpose to give rise to neuroblasts, or type-A cells. Neuroblasts, upon their generation, migrate away from the niche, following the rostral migratory stream, colonize the olfactory bulbs and differentiate into olfactory inhibitory interneurons.16,17 Multiciliated ependymal cell play essential role in the assembly of the SEZ niche, while they facilitate the cerebrospinal fluid (CSF) flow and they regulate the fate decisions of aNSCs within the SEZ. Multiciliated ependymal cells are organized in a monolayer in the ventricular surface and thus they form a barrier between the CSF and the adult SEZ niche.18 Ependymal cells have a unique morphology, which characterizes all the multiciliated cells in different tissues and organisms, as they carry in their apical surface multiple basal bodies from which motile cilia arise.19 Motile cilia, which are floating in the CSF, can receive secreted molecules and morphogens from the CSF and thus they influence aNSCs function. Additionally, through the co-ordinating movement of their motile cilia, ependymal cells regulate the CSF movement through the ventricular system. By this movement they help in the maintenance of the differential distribution of the molecules in the CSF resulting in different availability of these molecules to the SEZ niche stem cells across the rostro-caudal and dorso-ventral part of the niche, which in turn affects the behavior of aNSCs.18,20,21 Despite their involvement in the cell fate decisions of aNSCs, it was recently proposed that multiciliated ependymal cells may have a more active role in the adult SEZ niche. It was shown that, although they are postmitotic,18 they can be activated upon insult and generate neuroblasts and astrocytes.22
Recent work shed light on the molecular cascade generating multiciliated ependymal cells.13,18,23-26 Both type-B1 cells as well as multiciliated ependymal cells are generated from radial glial cell which, at the end of embryogenesis, have not yet been differentiated into neurons or glial cells. Ependymal cells are generated during the first 2 postnatal weeks from radial glial cells.18,23 This subpopulation of radial glial cells is committed to ependymal cell lineage before birth, at around E16 days post coitum (dpc), when they stop to proliferate but maintain their undifferentiated state until after birth.18 Foxj1 is a key factor in the initiation of radial glial cells differentiation during postnatal stages, as its activation is responsible for the docking of basal bodies into the apical surface of the newly generated ependymal cells,27,28 which is essential for the generation of multiple motile cilia. The differentiation of radial glial cells toward ependymal cells is an important step for the initiation of the assembly of the adult SEZ niche. The molecular pathway, which mediates this process involves, in addition to Foxj1, the Ank3 protein, a direct target of Foxj1, responsible for the integrity of the rosette-like structures, formed by ependymal cells and type-B1 cells, in the wall of the lateral ventricles.29 Thus, Foxj1 activation is necessary for radial glial cells differentiation, however little is known about the control of its expression. Recently it was shown that c-Myb acts upstream of Foxj1 regulating directly its expression in the multiciliation process of the mouse airway epithelium.30 In the same work it was shown that Mcidas activates c-Myb and Foxj1 in the context of mouse airway epithelium,30 but whether this is a general pathway in all multiciliated cells and whether other molecules are acting upstream of c-Myb and Foxj1 was not known until recently.
We have proposed that Mcidas and GemC1/Lynkeas are involved in the molecular pathway which controls radial glial cells fate commitment and multiciliated ependymal cells differentiation through both c-Myb and Foxj1 in the development of mouse brain.13 Mcidas and GemC1/Lynkeas expression is initiated in radial glial cells located at the periventricular zone at E16 dpc. Their expression is maintained until the first postnatal stages in the wall of the lateral ventricles and initially precedes that of FoxJ1, while at later stages they are colocalized with Foxj1.13 Mcidas protein is also colocalized with S100β, a marker of multiciliated ependymal cells, but not with Tuj1, marker of neurons, suggesting that Mcidas and GemC1/Lynkeas characterize radial glial cells committed to the ependymal cell lineage and thus they are important for radial glial cells specification toward this lineage. These new evidences make it possible to characterize the subtype of radial glial cells which gives rise to ependymal cells and reinforce previous data suggesting that radial glial cells which will differentiate into ependymal cells are committed to this lineage during embryogenesis, long before their differentiation.18
The role of Mcidas and GemC1/Lynkeas in radial glial cells fate commitment decisions was addressed by overexpression and depletion experiments.13 Their overexpression resulted in loss of radial glial characteristics of periventricular zone cells, failure in the migration of radial glial cells toward the cortical layers and failure in their subsequent differentiation into cortical neurons. In this region they differentiated into multiciliated ependymal cells as they generate multiple basal bodies and cilia. On the other hand depletion of Mcidas or GemC1/Lynkeas lead to the abolishment of c-Myb and Foxj1 expression leading to blockage of ependymal cells generation. These results indicate that both Mcidas and GemC1/Lynkeas are sufficient and necessary for radial glial cells fate commitment and multiciliated ependymal cells differentiation. Moreover, we have shown that Mcidas and GemC1/Lynkeas act upstream of c-Myb and FoxJ1 promoting their transcriptional activation. Upregulation of c-Myb is necessary for GemC1/Lynkeas and Mcidas to initiate ependymal cell fate as c-Myb depletion abolish the effects triggered by GemC1/Lynkeas and Mcidas overexpression, suggesting that the activation of c-Myb by GemC1/Lynkeas and Mcidas is a critical step for ependymal cell generation. Moreover, we have shown that GemC1/Lynkeas is responsible for the transcriptional activation of Mcidas expression, while Mcidas is also able to regulate its own expression13,31 (Fig. 1).
Figure 1.
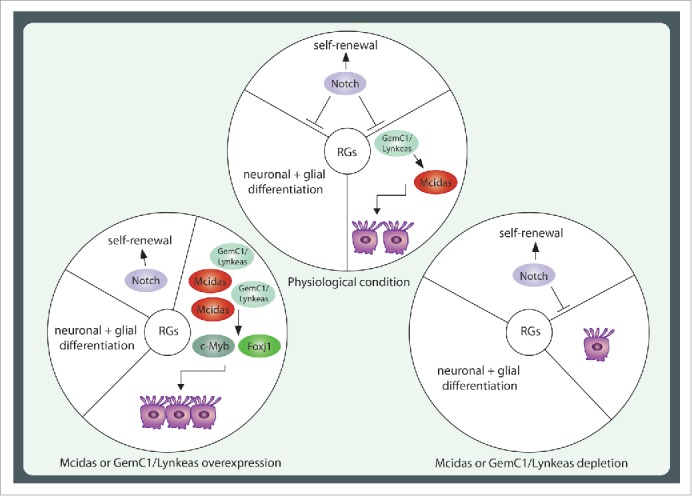
Schematic representation of Mcidas and GemC1/Lynkeas role in radial glial cells fate commitment decisions. (A) Radial glial cells under physiological conditions have the ability to perform self-renewal divisions at early developmental stages, while at later stages they differentiate toward neuronal and glial cells. In addition, radial glial cells generate the adult SEZ niche cells, the multiciliated ependymal cells and the type-B1 adult neural stem cells. Mcidas and GemC1/Lynkeas control the molecular pathway responsible for the generation of multiciliated ependymal cells, acting antagonistically to Notch signaling, which maintains radial glial cells in undifferentiated state and acting upstream of c-Myb and Foxj1, known for their role in multiciliated cell differentiation. (B) Upon Mcidas and GemC1/Lynkeas overexpression there is a shift of radial glial cells fate decisions toward the multiciliated ependymal cell lineage, as the molecular pathway, involving c-Myb and Foxj1 is initiated earlier. (C) Upon Mcidas or GemC1/Lynkeas depletion, the commitment of radial glial cell toward the ependymal cell lineage is blocked, resulting in decrease of c-Myb and Foxj1 expressing cells and leads consequently to a decrease of the multiciliated ependymal cells generated in the wall of the lateral ventricles. In addition, their depletion leads to increased number of GFAP+ astroglial or astroglial-like cells suggesting a change in the fate commitment decisions of radial glial cells. RGs: radial glial cells.
The molecular pathway activated by Mcidas and GemC1/Lynkeas is negatively regulated by Notch signaling, which maintains radial glial cells in an undifferentiated state by antagonizing GemC1/Lynkeas and Mcidas.13 This is of great interest as only a small subpopulation of radial glial cells will be differentiated into multiciliated ependymal cells and therefore the antagonistic activity of Notch signaling on the one side and GemC1/Lynkeas and Mcidas on the other could explain how radial glial cells fate decisions are regulated during mouse brain development (Fig. 1).
Concluding we have shown for the first time that GemC1/Lynkeas and Mcidas are key components of the molecular cascade regulating multiciliated ependymal cells generation. They act high in the hierarchy, upstream of c-Myb and Foxj1, by activating their expression, while they antagonise Notch signaling. Additionally we have provided evidence on the functional hierarchy between the 2 genes with GemC1/Lynkeas to transcriptionally activate Mcidas expression. Due to their temporal expression, it is plausible that GemC1/Lynkeas initiates the fate specification of radial glial cells, while Mcidas is playing a more active role on the establishment and maintenance of the multiciliated ependymal cell fate. Key events on this pathway are the activation of c-Myb and FoxJ1 which are responsible for the full differentiation of these cells. In agreement with this idea GemC1/Lynkeas and Mcidas expression is reduced postnatally prior to the generation of fully differentiated ependymal cells. The maintenance of Foxj1 expression at postnatal stages could be responsible for the propagation of the differentiation of multiciliated ependymal cells, which is only taking place at postnatal stages.
The molecular pathway of GemC1/Lynkeas-Mcidas-c-Myb-Foxj1 is highly conserved on different tissues and among different organisms, as we have shown that GemC1/Lynkeas also participates in the differentiation of multiciliated cells in the mouse airway epithelium,31 while GemC1/Lynkeas also controls the multiciliation process in zebrafish.32 Taking into account these results and given the fact that Mcidas is essential for multiciliation process in human airway,31,33 it is plausible that GemC1/Lynkeas also participates in multicilated cell differentitaion in humans. GemC1/Lynkeas might be essential in multiciliated ependymal cells generation in the adult human brain and therefore could be a target molecule for understanding how the adult human SEZ niche is assembled and functions.
Abbreviations
- aNSCs
Adult neural stem cells
- CSF
cerebrospinal fluid
- Dpc
days post coitum
- SEZ
subependymal zone
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all members of our laboratories for helpful discussions and for their comments on the manuscript and Meletios Verras for the illustrations.
Funding
This work was supported by Thallis research program “The role and the mechanisms that control the asymmetric cell division during stem cells differentiation,” European Research Council (ERC, DYNACOM 281851) and Aristeia II, GEMCCTR “Self-renewal and differentiation decisions in neural stem cells: Geminin, cell cycle control and transcriptional regulation.”
References
- [1].Caillat C, Fish A, Pefani DE, Taraviras S, Lygerou Z, Perrakis A. The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled-coil proteins. Acta Crystallogr D Biol Crystallogr 2015; 71:2278-86; PMID:26527144; http://dx.doi.org/ 10.1107/S1399004715016892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Caillat C, Pefani DE, Gillespie PJ, Taraviras S, Blow JJ, Lygerou Z, Perrakis A. The Geminin and Idas coiled coils preferentially form a heterodimer that inhibits Geminin function in DNA replication licensing. J Biol Chem 2013; 288:31624-34; PMID:24064211; http://dx.doi.org/ 10.1074/jbc.M113.491928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pefani DE, Dimaki M, Spella M, Karantzelis N, Mitsiki E, Kyrousi C, Symeonidou IE, Perrakis A, Taraviras S, Lygerou Z. Idas, a novel phylogenetically conserved geminin-related protein, binds to geminin and is required for cell cycle progression. J Biol Chem 2011; 286:23234-46; PMID:21543332; http://dx.doi.org/ 10.1074/jbc.M110.207688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol 2010; 12:484-91; PMID:20383140; http://dx.doi.org/ 10.1038/ncb2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Champeris Tsaniras S, Kanellakis N, Symeonidou IE, Nikolopoulou P, Lygerou Z, Taraviras S. Licensing of DNA replication, cancer, pluripotency and differentiation: an interlinked world? Semin Cell Dev Biol 2014; 30:174-80; PMID:24641889; http://dx.doi.org/ 10.1016/j.semcdb.2014.03.013 [DOI] [PubMed] [Google Scholar]
- [6].Symeonidou IE, Taraviras S, Lygerou Z. Control over DNA replication in time and space. FEBS Lett 2012; 586:2803-12; PMID:22841721; http://dx.doi.org/ 10.1016/j.febslet.2012.07.042 [DOI] [PubMed] [Google Scholar]
- [7].Spella M, Britz O, Kotantaki P, Lygerou Z, Nishitani H, Ramsay RG, Flordellis C, Guillemot F, Mantamadiotis T, Taraviras S. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience 2007; 147:373-87; PMID:17533120; http://dx.doi.org/ 10.1016/j.neuroscience.2007.03.050 [DOI] [PubMed] [Google Scholar]
- [8].Spella M, Kyrousi C, Kritikou E, Stathopoulou A, Guillemot F, Kioussis D, Pachnis V, Lygerou Z, Taraviras S. Geminin regulates cortical progenitor proliferation and differentiation. Stem Cells 2011; 29:1269-82; PMID:21681860; http://dx.doi.org/ 10.1002/stem.678 [DOI] [PubMed] [Google Scholar]
- [9].Yellajoshyula D, Lim JW, Thompson DM Jr., Witt JS, Patterson ES, Kroll KL. Geminin regulates the transcriptional and epigenetic status of neuronal fate-promoting genes during mammalian neurogenesis. Mol Cell Biol 2012; 32:4549-60; PMID:22949506; http://dx.doi.org/ 10.1128/MCB.00737-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Karamitros D, Patmanidi AL, Kotantaki P, Potocnik AJ, Bahr-Ivacevic T, Benes V, Lygerou Z, Kioussis D, Taraviras S. Geminin deletion increases the number of fetal hematopoietic stem cells by affecting the expression of key transcription factors. Development 2015; 142:70-81; PMID:25516969; http://dx.doi.org/ 10.1242/dev.109454 [DOI] [PubMed] [Google Scholar]
- [11].Roukos V, Iliou MS, Nishitani H, Gentzel M, Wilm M, Taraviras S, Lygerou Z. Geminin cleavage during apoptosis by caspase-3 alters its binding ability to the SWI/SNF subunit Brahma. J Biol Chem 2007; 282:9346-57; PMID:17261582; http://dx.doi.org/ 10.1074/jbc.M611643200 [DOI] [PubMed] [Google Scholar]
- [12].Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev 2005; 19:1723-34; PMID:16024661; http://dx.doi.org/ 10.1101/gad.1319105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kyrousi C, Arbi M, Pilz GA, Pefani DE, Lalioti ME, Ninkovic J, Götz M, Lygerou Z, Taraviras S. Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development 2015; 142:3661-74; PMID:26395491; http://dx.doi.org/ 10.1242/dev.126342 [DOI] [PubMed] [Google Scholar]
- [14].Fuentealba LC, Obernier K, Alvarez-Buylla A. Adult neural stem cells bridge their niche. Cell Stem Cell 2012; 10:698-708; PMID:22704510; http://dx.doi.org/ 10.1016/j.stem.2012.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lim DA, Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci 2014; 37:563-71; PMID:25223700; http://dx.doi.org/ 10.1016/j.tins.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science 1994; 264:1145-8; PMID:8178174; http://dx.doi.org/ 10.1126/science.8178174 [DOI] [PubMed] [Google Scholar]
- [17].Carleton A, Petreanu LT, Lansford R, Alvarez-Buylla A, Lledo PM. Becoming a new neuron in the adult olfactory bulb. Nat Neurosci 2003; 6:507-18; PMID:12704391. [DOI] [PubMed] [Google Scholar]
- [18].Spassky N, Merkle FT, Flames N, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci 2005; 25:10-18; PMID:15634762; http://dx.doi.org/ 10.1523/JNEUROSCI.1108-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell 2008; 3:265-78; PMID:18786414; http://dx.doi.org/ 10.1016/j.stem.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JL, et al.. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science 2006; 311:629-32; PMID:16410488; http://dx.doi.org/ 10.1126/science.1119133 [DOI] [PubMed] [Google Scholar]
- [21].Mirzadeh Z, Han YG, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Cilia organize ependymal planar polarity. J Neurosci 2010; 30:2600-10; PMID:20164345; http://dx.doi.org/ 10.1523/JNEUROSCI.3744-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Carlen M, Meletis K, Goritz C, Darsalia V, Evergren E, Tanigaki K, Amendola M, Barnabé-Heider F, Yeung MS, Naldini L, et al.. Forebrain ependymal cells are Notch-dependent and generate neuroblasts and astrocytes after stroke. Nat Neurosci 2009; 12:259-67; PMID:19234458; http://dx.doi.org/ 10.1038/nn.2268 [DOI] [PubMed] [Google Scholar]
- [23].Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A 2004; 101:17528-32; PMID:15574494; http://dx.doi.org/ 10.1073/pnas.0407893101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Young KM, Fogarty M, Kessaris N, Richardson WD. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J Neurosci 2007; 27:8286-96; PMID:17670975; http://dx.doi.org/ 10.1523/JNEUROSCI.0476-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, Alvarez-Buylla A. Embryonic Origin of Postnatal Neural Stem Cells. Cell 2015; 161:1644-55; PMID:26091041; http://dx.doi.org/ 10.1016/j.cell.2015.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 2009; 136:4021-31; PMID:19906869; http://dx.doi.org/ 10.1242/dev.041129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 2008; 40:1454-60; PMID:19011629; http://dx.doi.org/ 10.1038/ng.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 2008; 40:1445-53; PMID:19011630; http://dx.doi.org/ 10.1038/ng.263 [DOI] [PubMed] [Google Scholar]
- [29].Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, Kuo CT. Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron 2011; 71:61-75; PMID:21745638; http://dx.doi.org/ 10.1016/j.neuron.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez-Buylla A, Stearns T, Kintner C, et al.. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development 2013; 140:4277-86; PMID:24048590; http://dx.doi.org/ 10.1242/dev.094102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Arbi M, Pefani DE, Kyrousi C, Lalioti ME, Kalogeropoulou A, Papanastasiou AD, Taraviras S, Lygerou Z. GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep 2016; 17(3):400-13; PMID:26882546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou F, Narasimhan V, Shboul M, Chong YL, Reversade B, Roy S. Gmnc Is a Master Regulator of the Multiciliated Cell Differentiation Program. Curr Biol 2015; 25(4):3267-73; http://dx.doi.org/ 10.1016/j.cub.2015.10.062 [DOI] [PubMed] [Google Scholar]
- [33].Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, et al.. MCIDAS mutations result in a mucociliary clearance disorder with reduced generation of multiple motile cilia. Nat Commun 2014; 5:4418; PMID:25048963; http://dx.doi.org/ 10.1038/ncomms5418 [DOI] [PubMed] [Google Scholar]
- [34].Patsi-Garin E. Dictionary of Greek Mythology. Athens: Patsiw Haris; 1969. [Google Scholar]


