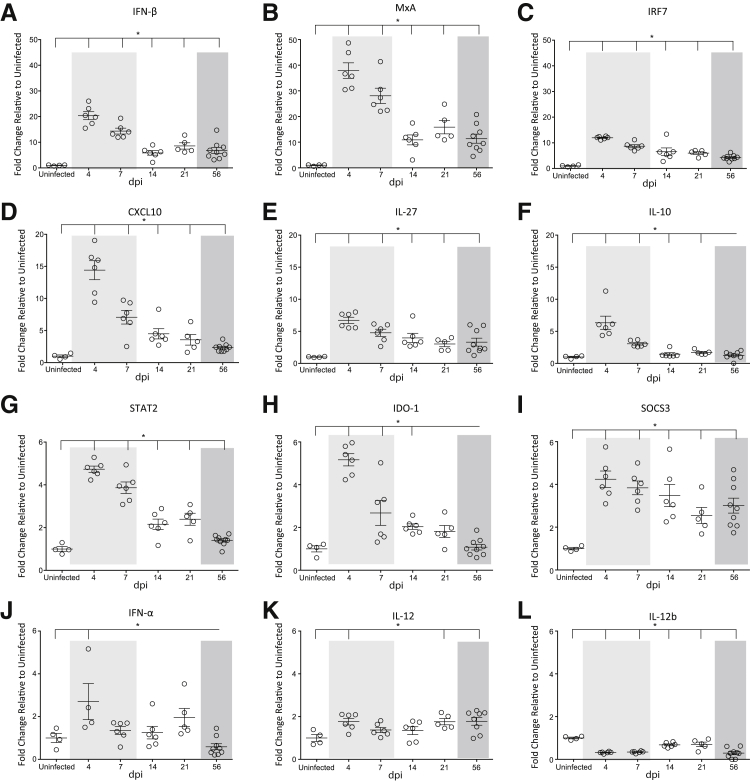
Figure 3.
Quantitation of interferon-stimulated genes. RNA was isolated from the spleen in uninfected animals and at 4, 7, 14, 21, and 56 dpi and gene expression analyses performed by NanoString barcoding technology. The relative counts of each gene product were determined on normalizing to four housekeeping genes. The fold-change of each gene was determined relative to uninfected controls, which was set to 1. The longitudinal fold-changes in interferon (IFN)-β (A), MxA (B), IRF7 (C), CXCL10 (D), IL-27 (E), IL-10 (F), STAT2 (G), IDO-1 (H), SOCS3 (I), IFN-α (J), IL-12 (K), and IL-12b (L) in the spleen. The acute (light shading), asymptomatic, and chronic (dark shading) phases of SIV infection are depicted. Statistical significance was determined by Kruskal-Wallis analysis of variance test followed by Dunn's multiple comparison analysis. Vertical lines indicate a significant difference between the indicated dpi and uninfected animals, as determined by the Dunn's multiple comparison analysis. Data are expressed as means ± SD. ∗P ≤ 0.05 for infected animals at all time points relative to uninfected macaques.