Figure 1.
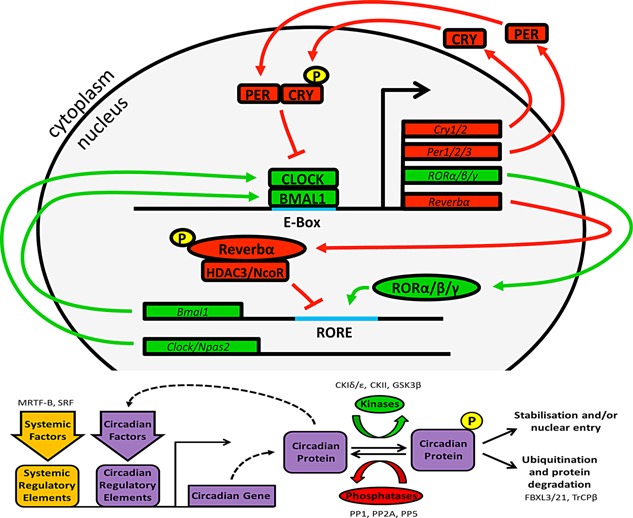
The molecular clockwork in mammals. In mammals, the molecular basis of circadian timing involves a transcriptional/translational/post‐translational feedback loop centred on the transcriptional activators CLOCK (or NPAS2) and BMAL1 and repressors PERIOD (PER1,2,3) and CRYPTOCHROME (CRY1,2). CLOCK and BMAL1 heterodimers bind to E‐box recognition sites to drive transcription of Period (Per1/2/3) and Cryptochrome (Cry1/2) 11, 12, 13, 14, 15, 16. PER and CRY proteins subsequently interact to form the core of a repressive complex, which inhibits BMAL1:CLOCK activity and expedites their clearance. Subsequent reduction of transcriptional activity at the Per and Cry loci, combined with active targeting of PER and CRY proteins for ubiquitination and degeneration (by TrCPβ and FXBL3/21, respectively) attenuates this repressive arm of the clock 116. CLOCK and BMAL1 concentrations rise once again to perpetuate the cycle. This core loop is influenced by auxiliary feedback loops, such as that involving the nuclear hormone receptors REV‐ERBα and RORα/β/γ 117, 118. Circadian proteins are also highly regulated by kinases and phosphatases which affect their stability, turnover, and sub‐cellular localization. Critically of course, components of the clock do not only drive their own expression cycles, but also impose rhythmic expression profiles onto a vast array of target genes through transcriptional enhancer element activity (e.g. via E‐Box and RORE elements), rhythmic chromatin modification (e.g. via CLOCK driven acetylation), and interaction with other non‐clock transcription factors (e.g. PER inhibition of PPARγ). In this way, transcriptional and biochemical processes within the cell are temporally coordinated across the cycle. Finally, many circadian factors are also responsive to systemic factors, which drive local cellular signaling pathways to achieve global synchronization of circadian phase.
