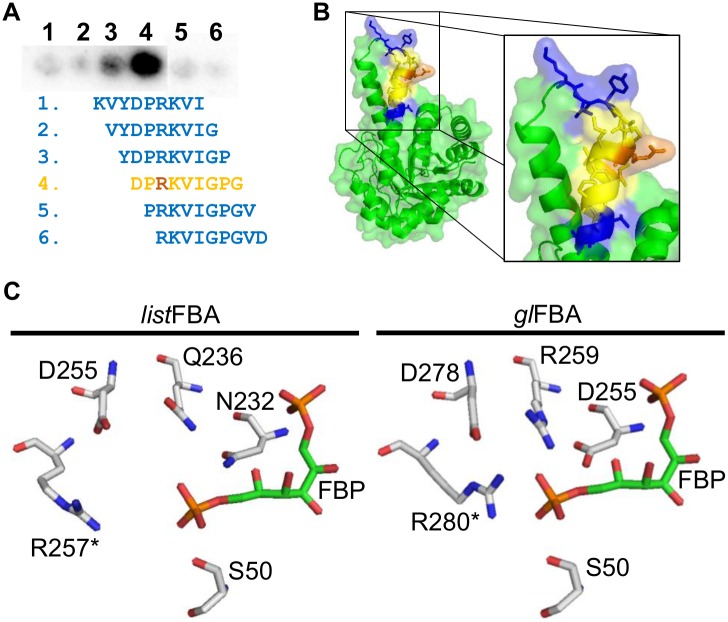
Fig 5. Epitope mapping of mAb-3F8 and position of the epitope in FBA structure.
(A) By using a peptide membrane, which was incubated with mAb-3F8 as primary antibody, it was possible to identify a 14-amino acid sequence as the epitope of mAb-3F8. In this sequence, it is also possible to observe that a 9-amino acid region (yellow) gives a higher reaction and, thus, is likely the most important recognition part of the epitope. Interestingly, Arg257 (orange), which may be part of the catalytic site of the dimeric form of FBA is present in the epitope. (B) The position of the epitope in FBA structure shows it is a loop-to-helix transition, which has many amino acid side chains exposed on the protein surface. (C) A closer look on the G3P site (part of the catalytic site) shows that Arg257 of Listeria spp. FBA (listFBA, left part) can play role on binding G3P, since binding pocket is similar to that from Giardia lamblia FBA (glFBA, right part). For graphical reasons, FBP molecule fitting in listFBA pocket has the same position of that one from glFBA, thus it may not represent the real fitting of this molecule in the structure. Asterisk (*) indicate that the amino acid is present in the partner subunit of the protein dimer.